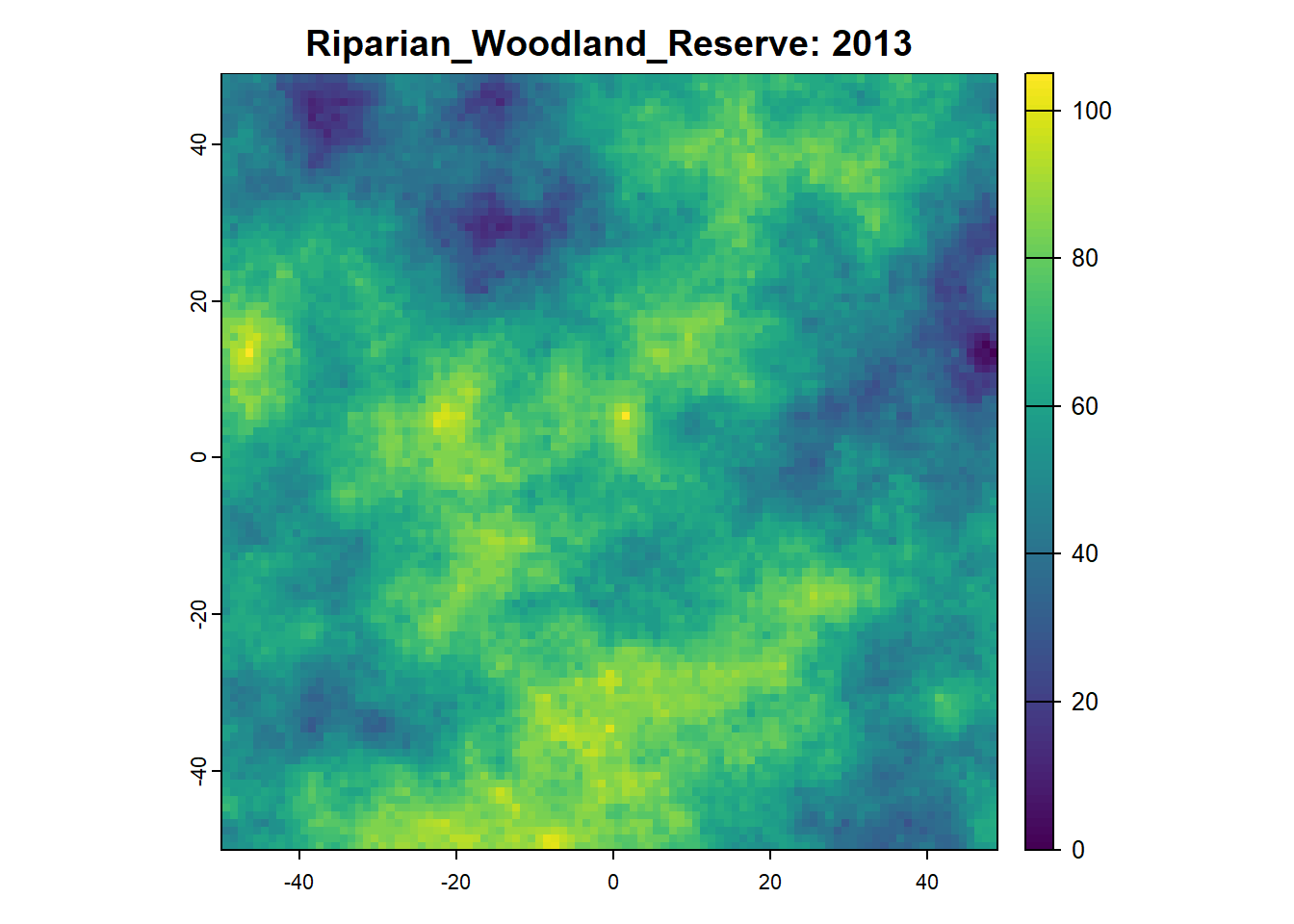
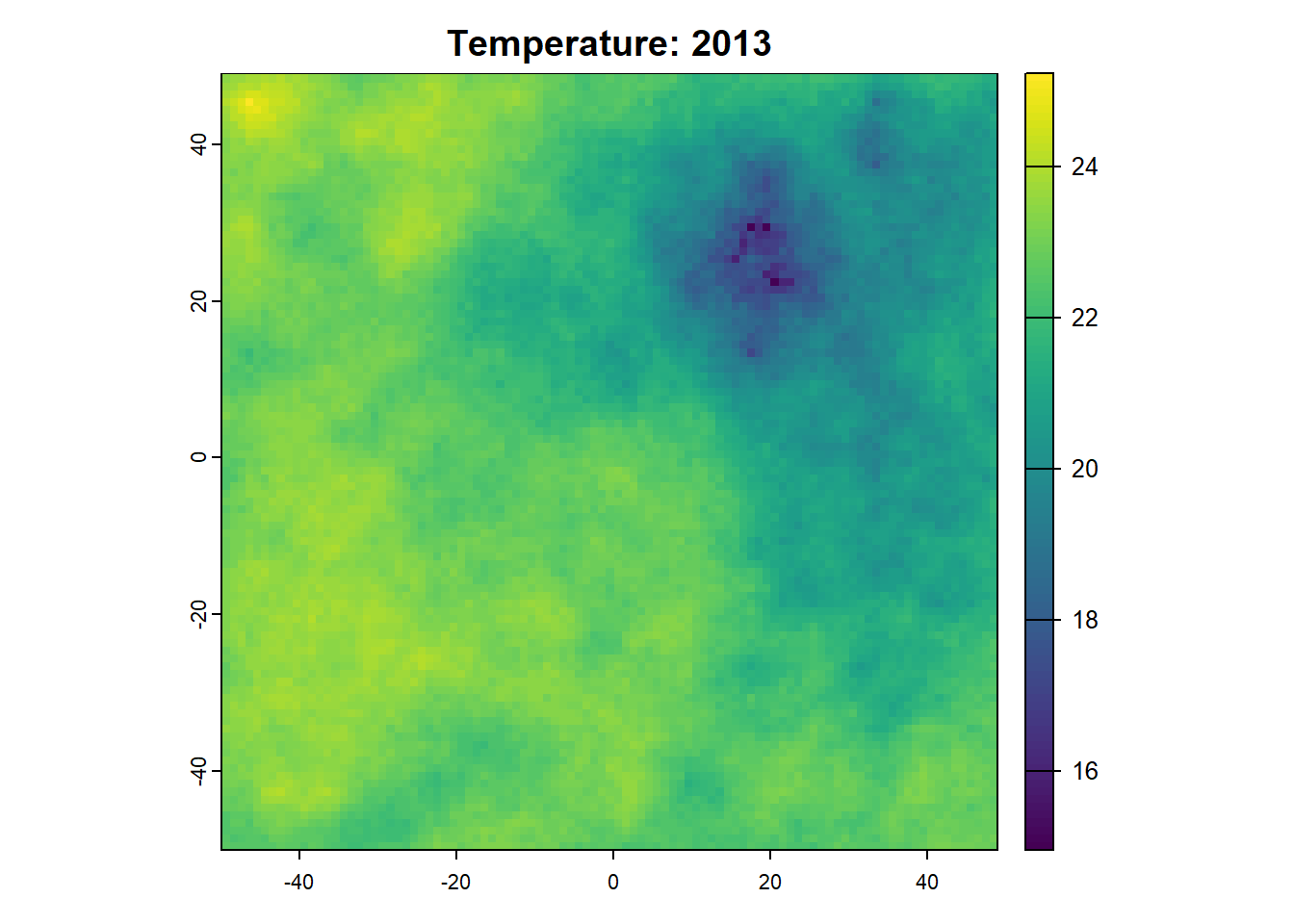
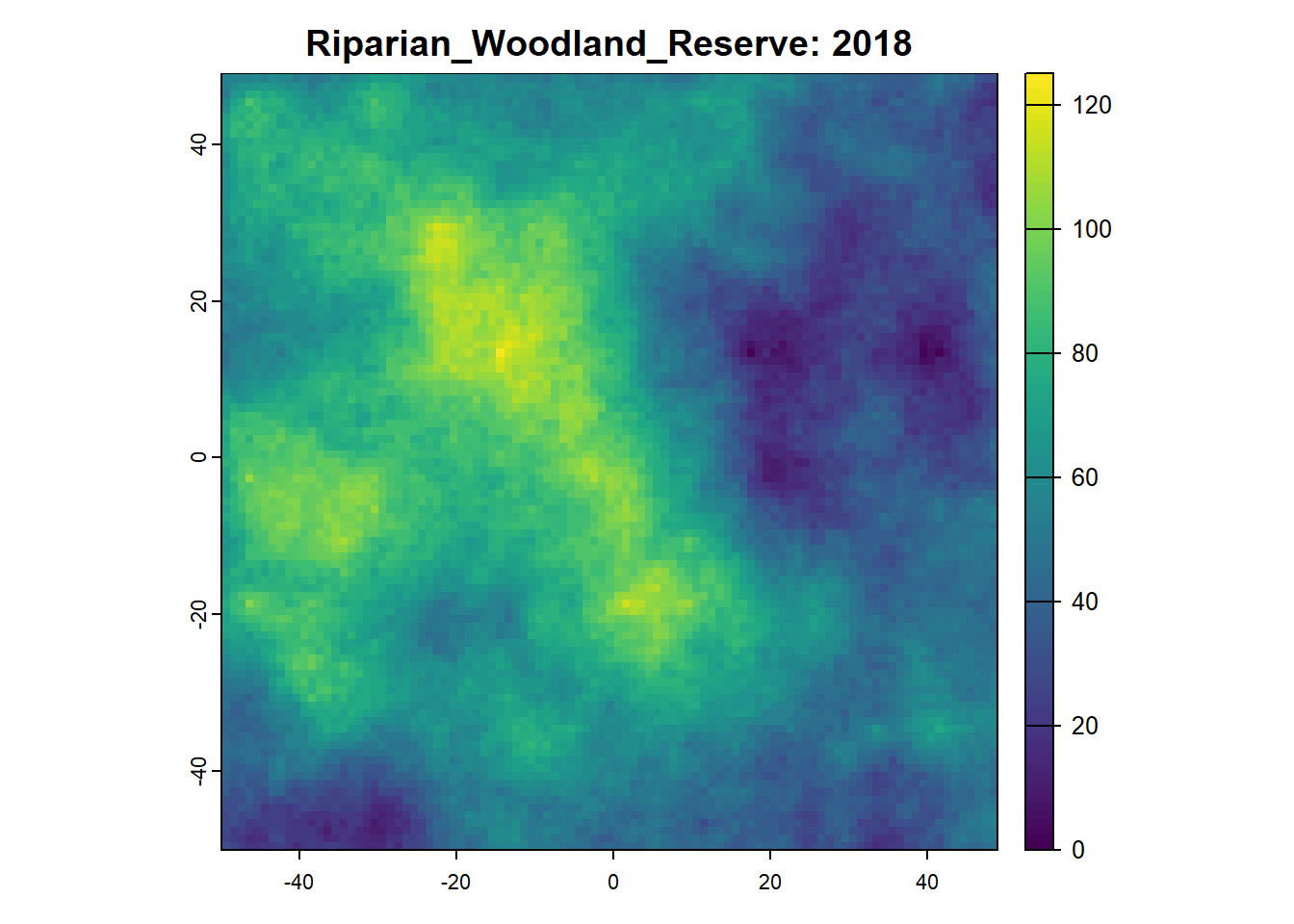
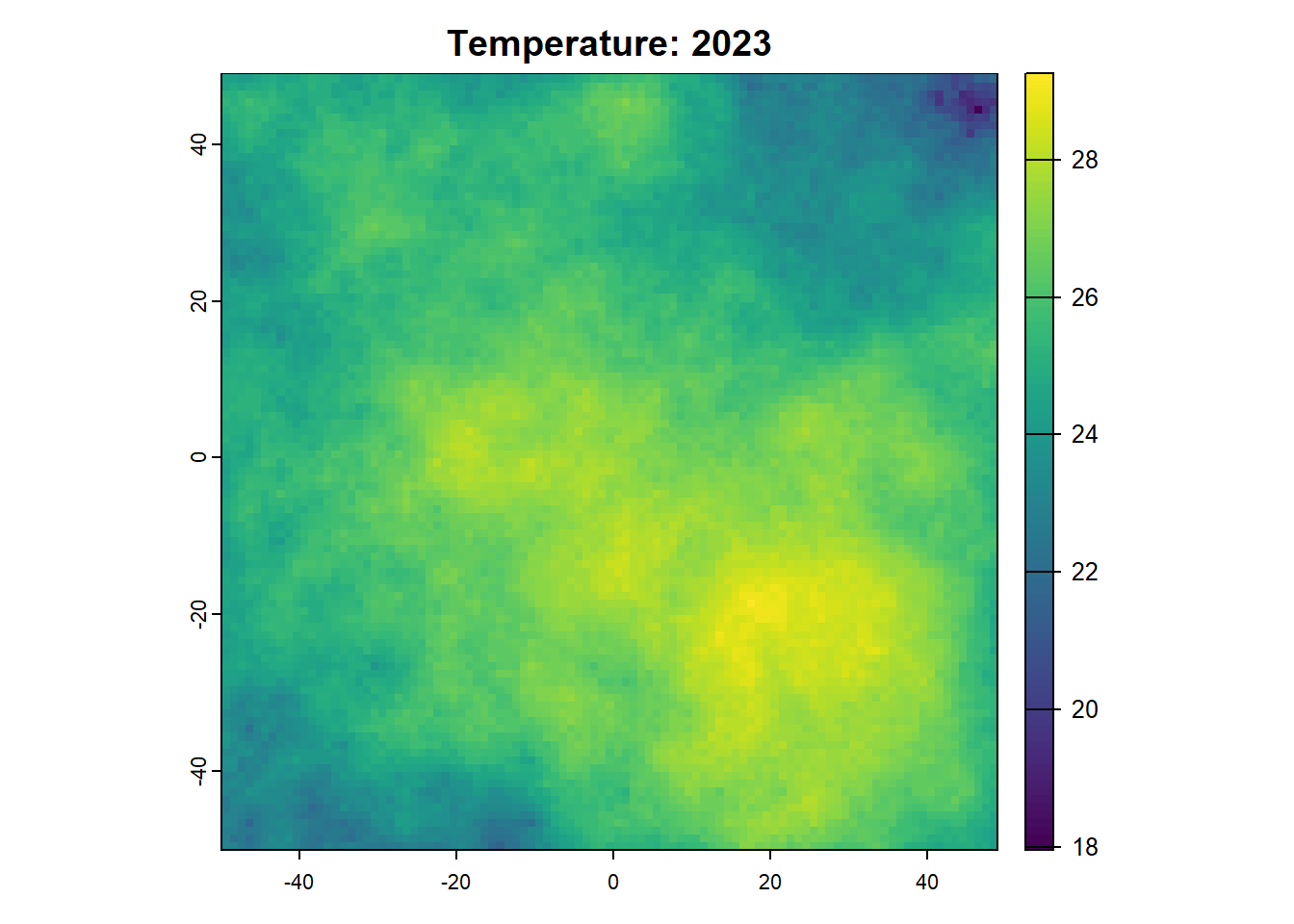
Once we want to run a project that is more sophisticated, we can start using SpaDES.project. SpaDES.projectSee here to see a longer list of project challenges .
We will demonstrate its use with an example created by Tati Micheletti where a workflow with several modules is setup and run.
SpaDES.project::setupProject
https://spades-project.predictiveecology.org/articles/i-getting-started.html
Installation
Code
<- c ("predictiveecology.r-universe.dev" , getOption ("repos" ))# if (!require("SpaDES.project")) :: Install (c ("SpaDES.project" , "SpaDES.core" , "reproducible" ), repos = repos, dependencies = TRUE )= SpaDES.project:: .libPathDefault ("~/SpaDES_book/integratingSpaDESmodules" )
Code
= SpaDES.project:: .libPathDefault ("PredictiveEcology.org" )
Using setupProject
setupProject does a series of very specific things, in a particular order, so that the chances of package conflicts and user errors are minimized.
Note how it downloads the listed modules directly from their GitHub repositories (e.g. tati-micheletti/speciesAbundance )
Code
##################### PART II: Download the modules and install the needed packages <- SpaDES.project:: setupProject (paths = list (projectPath = "~/SpaDES_book/integratingSpaDESmodules" ,packagePath = packagePath),modules = c ("tati-micheletti/speciesAbundance@main" ,"tati-micheletti/temperature@main" ,"tati-micheletti/speciesAbundTempLM@main" ),times = list (start = 2013 ,end = 2030 ),updateRprofile = FALSE
The output of setupProject is a list of argument values that can be passed to simInitAndSpaDES2 (with the “2” it accepts a list of arguments, instead of the arguments themselves).
Code
<- SpaDES.core:: simInitAndSpades2 (Setup)
Emergent workflow
We can evaluate the sequence of events using completed
Code
:: completed (results)
eventTime moduleName eventType eventPriority
<num> <char> <char> <num>
1: 2013 checkpoint init 0
2: 2013 save init 0
3: 2013 progress init 0
4: 2013 load init 0
5: 2013 speciesAbundance init 1
6: 2013 temperature init 1
7: 2013 speciesAbundTempLM init 1
8: 2013 speciesAbundance tableToRasters 5
9: 2013 speciesAbundance plot 5
10: 2013 temperature dataToRaster 5
11: 2013 temperature plotting 5
12: 2013 speciesAbundTempLM tableBuilding 5
13: 2014 speciesAbundance tableToRasters 5
14: 2014 temperature dataToRaster 5
15: 2014 speciesAbundTempLM tableBuilding 5
16: 2015 speciesAbundance tableToRasters 5
17: 2015 temperature dataToRaster 5
18: 2015 speciesAbundTempLM tableBuilding 5
19: 2016 speciesAbundance tableToRasters 5
20: 2016 temperature dataToRaster 5
21: 2016 speciesAbundTempLM tableBuilding 5
22: 2017 speciesAbundance tableToRasters 5
23: 2017 temperature dataToRaster 5
24: 2017 speciesAbundTempLM tableBuilding 5
25: 2018 speciesAbundance plot 5
26: 2018 speciesAbundance tableToRasters 5
27: 2018 temperature dataToRaster 5
28: 2018 speciesAbundTempLM tableBuilding 5
29: 2019 speciesAbundance tableToRasters 5
30: 2019 temperature dataToRaster 5
31: 2019 speciesAbundTempLM tableBuilding 5
32: 2020 speciesAbundance tableToRasters 5
33: 2020 temperature dataToRaster 5
34: 2020 speciesAbundTempLM tableBuilding 5
35: 2021 speciesAbundance tableToRasters 5
36: 2021 temperature dataToRaster 5
37: 2021 speciesAbundTempLM tableBuilding 5
38: 2022 speciesAbundance tableToRasters 5
39: 2022 temperature dataToRaster 5
40: 2022 speciesAbundTempLM tableBuilding 5
41: 2023 temperature plotting 5
42: 2023 temperature dataToRaster 5
43: 2023 speciesAbundTempLM tableBuilding 5
44: 2023 speciesAbundTempLM modelBuilding 5
45: 2023 speciesAbundTempLM abundanceForecasting 5
46: 2023 speciesAbundTempLM plot 5
47: 2024 temperature dataToRaster 5
48: 2024 speciesAbundTempLM abundanceForecasting 5
49: 2024 speciesAbundTempLM plot 5
50: 2025 temperature dataToRaster 5
51: 2025 speciesAbundTempLM abundanceForecasting 5
52: 2025 speciesAbundTempLM plot 5
53: 2026 temperature dataToRaster 5
54: 2026 speciesAbundTempLM abundanceForecasting 5
55: 2026 speciesAbundTempLM plot 5
56: 2027 temperature dataToRaster 5
57: 2027 speciesAbundTempLM abundanceForecasting 5
58: 2027 speciesAbundTempLM plot 5
59: 2028 temperature dataToRaster 5
60: 2028 speciesAbundTempLM abundanceForecasting 5
61: 2028 speciesAbundTempLM plot 5
62: 2029 temperature dataToRaster 5
63: 2029 speciesAbundTempLM abundanceForecasting 5
64: 2029 speciesAbundTempLM plot 5
65: 2030 temperature dataToRaster 5
66: 2030 speciesAbundTempLM abundanceForecasting 5
67: 2030 speciesAbundTempLM plot 5
eventTime moduleName eventType eventPriority
._prevEventTimeFinish clockTime
<POSc> <POSc>
1: 2024-09-18 08:36:00 2024-09-18 08:36:00
2: 2024-09-18 08:36:00 2024-09-18 08:36:00
3: 2024-09-18 08:36:00 2024-09-18 08:36:00
4: 2024-09-18 08:36:00 2024-09-18 08:36:00
5: 2024-09-18 08:36:00 2024-09-18 08:36:00
6: 2024-09-18 08:36:00 2024-09-18 08:36:00
7: 2024-09-18 08:36:00 2024-09-18 08:36:00
8: 2024-09-18 08:36:00 2024-09-18 08:36:00
9: 2024-09-18 08:36:00 2024-09-18 08:36:00
10: 2024-09-18 08:36:00 2024-09-18 08:36:00
11: 2024-09-18 08:36:00 2024-09-18 08:36:00
12: 2024-09-18 08:36:00 2024-09-18 08:36:00
13: 2024-09-18 08:36:00 2024-09-18 08:36:00
14: 2024-09-18 08:36:00 2024-09-18 08:36:00
15: 2024-09-18 08:36:00 2024-09-18 08:36:00
16: 2024-09-18 08:36:00 2024-09-18 08:36:00
17: 2024-09-18 08:36:00 2024-09-18 08:36:00
18: 2024-09-18 08:36:00 2024-09-18 08:36:00
19: 2024-09-18 08:36:00 2024-09-18 08:36:01
20: 2024-09-18 08:36:01 2024-09-18 08:36:01
21: 2024-09-18 08:36:01 2024-09-18 08:36:01
22: 2024-09-18 08:36:01 2024-09-18 08:36:01
23: 2024-09-18 08:36:01 2024-09-18 08:36:01
24: 2024-09-18 08:36:01 2024-09-18 08:36:01
25: 2024-09-18 08:36:01 2024-09-18 08:36:01
26: 2024-09-18 08:36:01 2024-09-18 08:36:01
27: 2024-09-18 08:36:01 2024-09-18 08:36:01
28: 2024-09-18 08:36:01 2024-09-18 08:36:01
29: 2024-09-18 08:36:01 2024-09-18 08:36:01
30: 2024-09-18 08:36:01 2024-09-18 08:36:01
31: 2024-09-18 08:36:01 2024-09-18 08:36:01
32: 2024-09-18 08:36:01 2024-09-18 08:36:01
33: 2024-09-18 08:36:01 2024-09-18 08:36:01
34: 2024-09-18 08:36:01 2024-09-18 08:36:01
35: 2024-09-18 08:36:01 2024-09-18 08:36:01
36: 2024-09-18 08:36:01 2024-09-18 08:36:01
37: 2024-09-18 08:36:01 2024-09-18 08:36:01
38: 2024-09-18 08:36:01 2024-09-18 08:36:01
39: 2024-09-18 08:36:01 2024-09-18 08:36:01
40: 2024-09-18 08:36:01 2024-09-18 08:36:01
41: 2024-09-18 08:36:01 2024-09-18 08:36:01
42: 2024-09-18 08:36:01 2024-09-18 08:36:01
43: 2024-09-18 08:36:01 2024-09-18 08:36:01
44: 2024-09-18 08:36:01 2024-09-18 08:36:01
45: 2024-09-18 08:36:01 2024-09-18 08:36:01
46: 2024-09-18 08:36:01 2024-09-18 08:36:01
47: 2024-09-18 08:36:01 2024-09-18 08:36:01
48: 2024-09-18 08:36:01 2024-09-18 08:36:01
49: 2024-09-18 08:36:01 2024-09-18 08:36:01
50: 2024-09-18 08:36:01 2024-09-18 08:36:01
51: 2024-09-18 08:36:01 2024-09-18 08:36:01
52: 2024-09-18 08:36:01 2024-09-18 08:36:02
53: 2024-09-18 08:36:02 2024-09-18 08:36:02
54: 2024-09-18 08:36:02 2024-09-18 08:36:02
55: 2024-09-18 08:36:02 2024-09-18 08:36:02
56: 2024-09-18 08:36:02 2024-09-18 08:36:02
57: 2024-09-18 08:36:02 2024-09-18 08:36:02
58: 2024-09-18 08:36:02 2024-09-18 08:36:02
59: 2024-09-18 08:36:02 2024-09-18 08:36:02
60: 2024-09-18 08:36:02 2024-09-18 08:36:02
61: 2024-09-18 08:36:02 2024-09-18 08:36:02
62: 2024-09-18 08:36:02 2024-09-18 08:36:02
63: 2024-09-18 08:36:02 2024-09-18 08:36:02
64: 2024-09-18 08:36:02 2024-09-18 08:36:02
65: 2024-09-18 08:36:02 2024-09-18 08:36:02
66: 2024-09-18 08:36:02 2024-09-18 08:36:02
67: 2024-09-18 08:36:02 2024-09-18 08:36:02
._prevEventTimeFinish clockTime
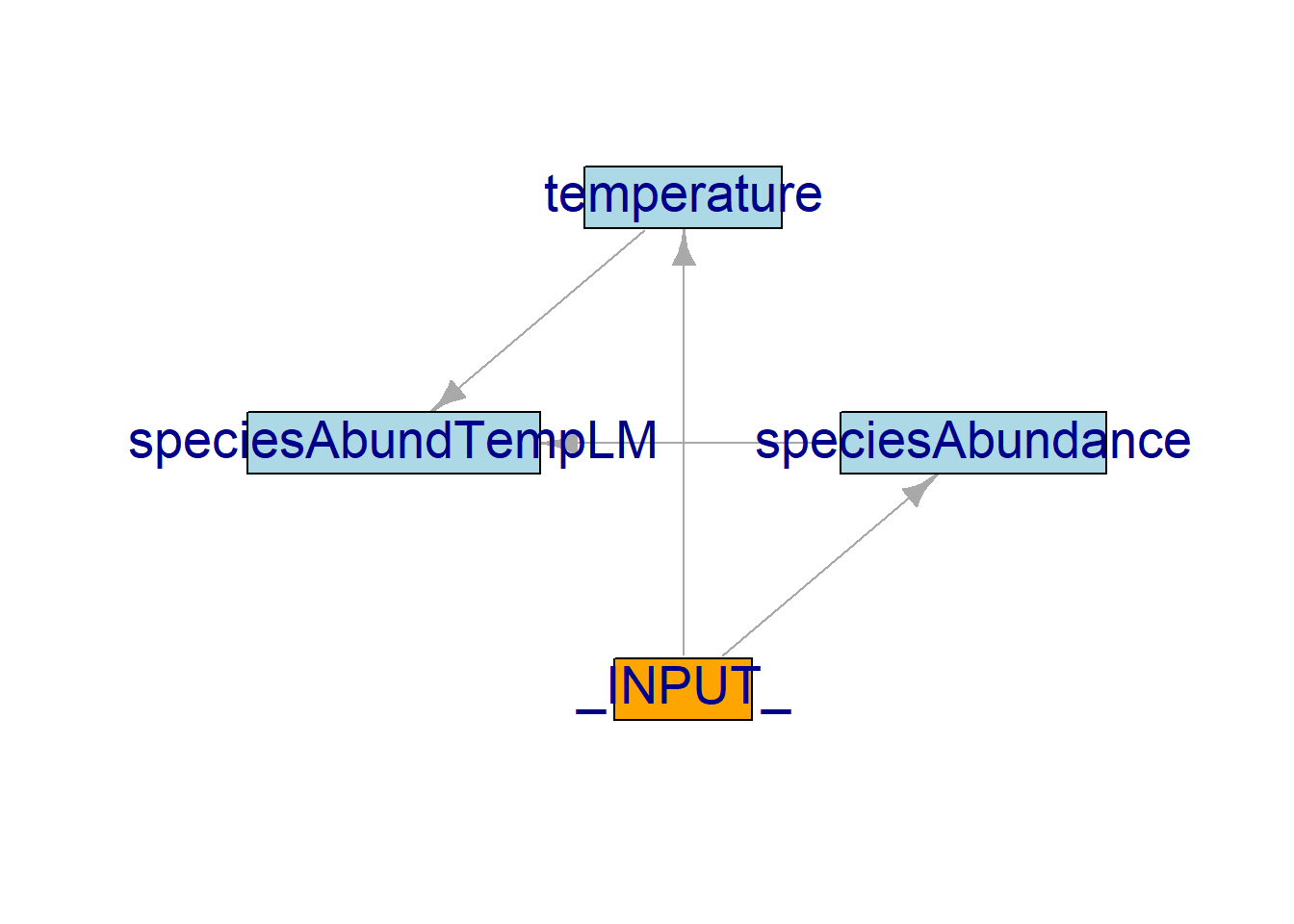
We can also see how modules and objects interact using the functions objectDiagram() and moduleDiagram().
Modules’ diagram showing the interactions among modules:
Code
:: moduleDiagram (results)
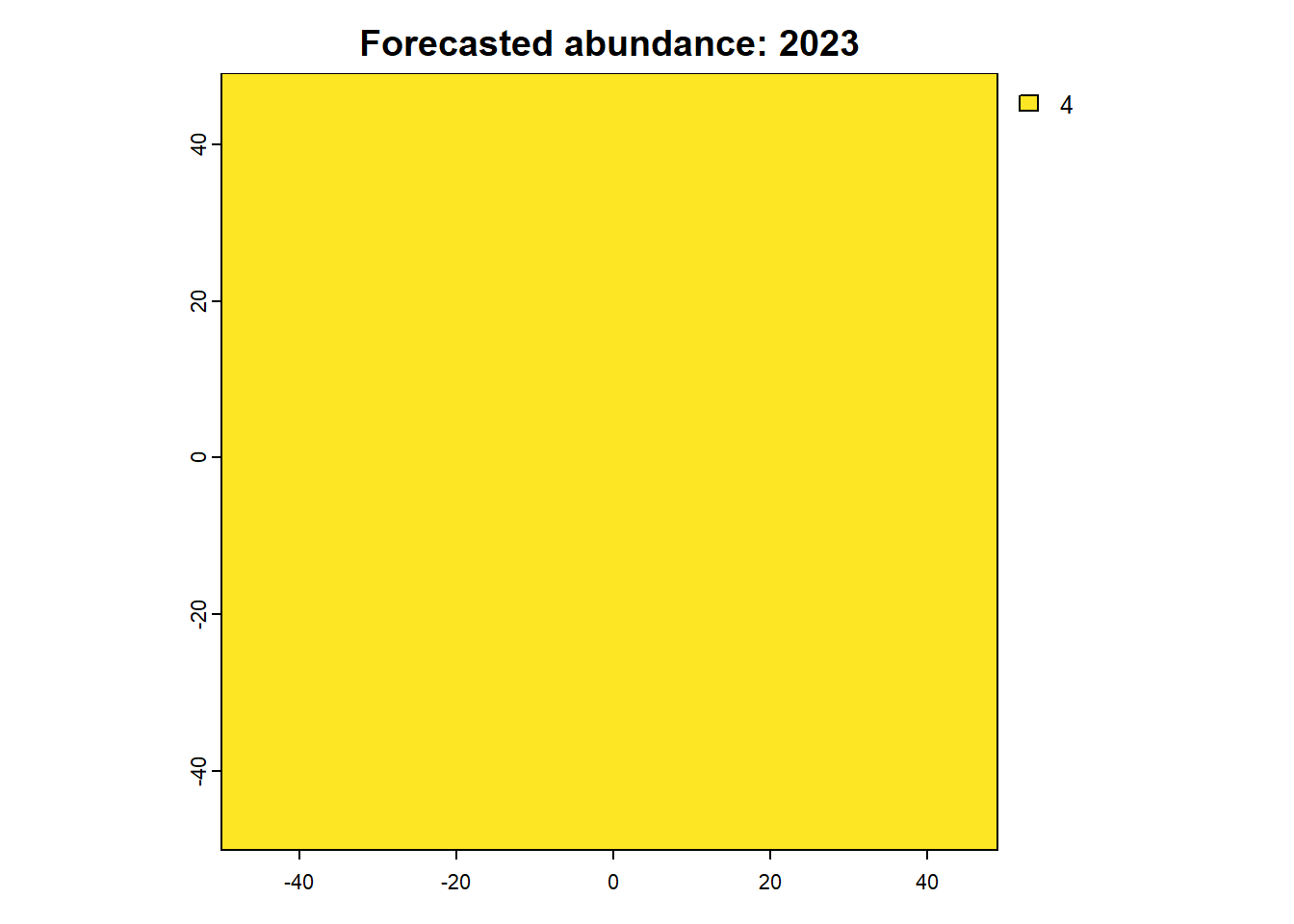
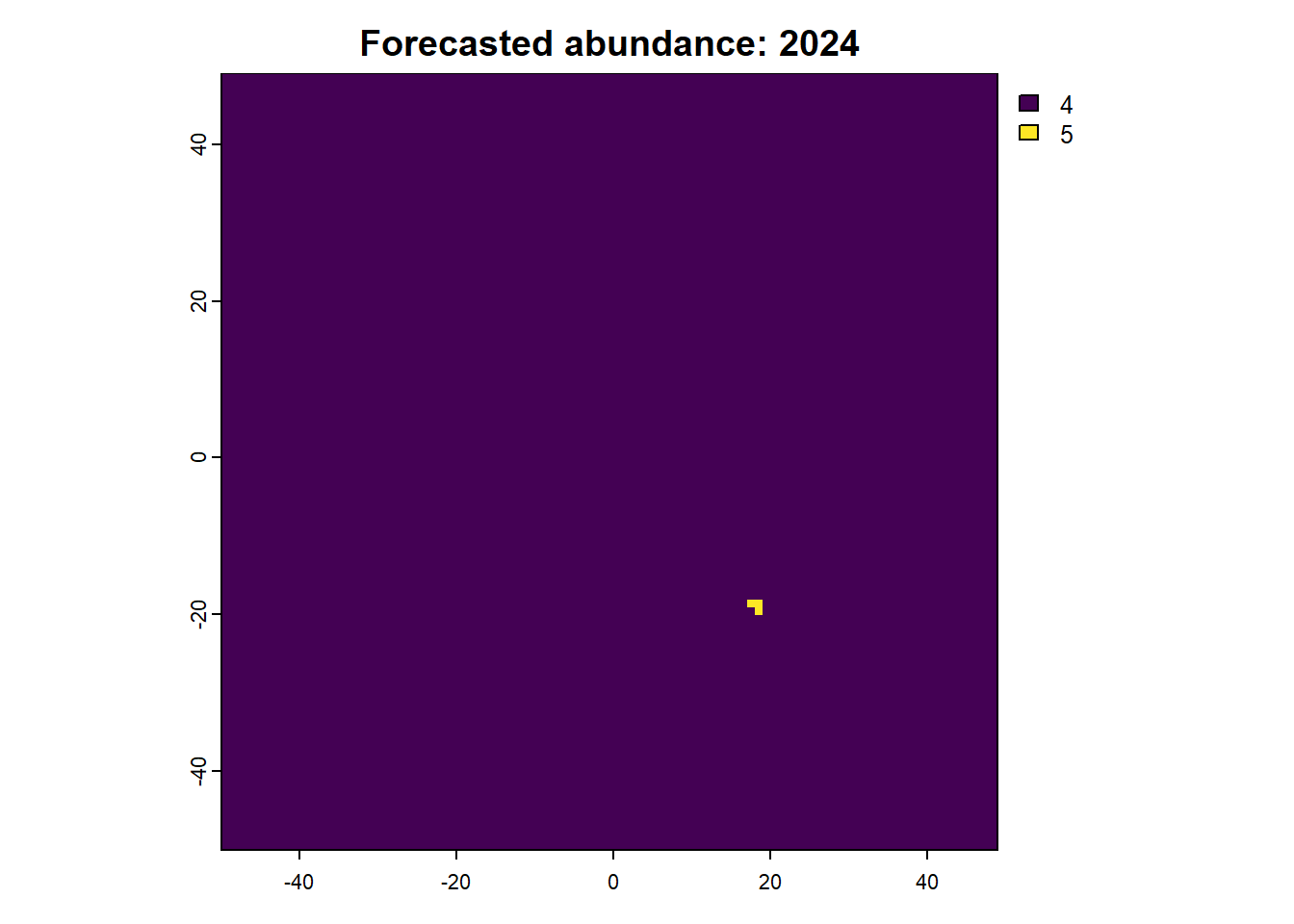
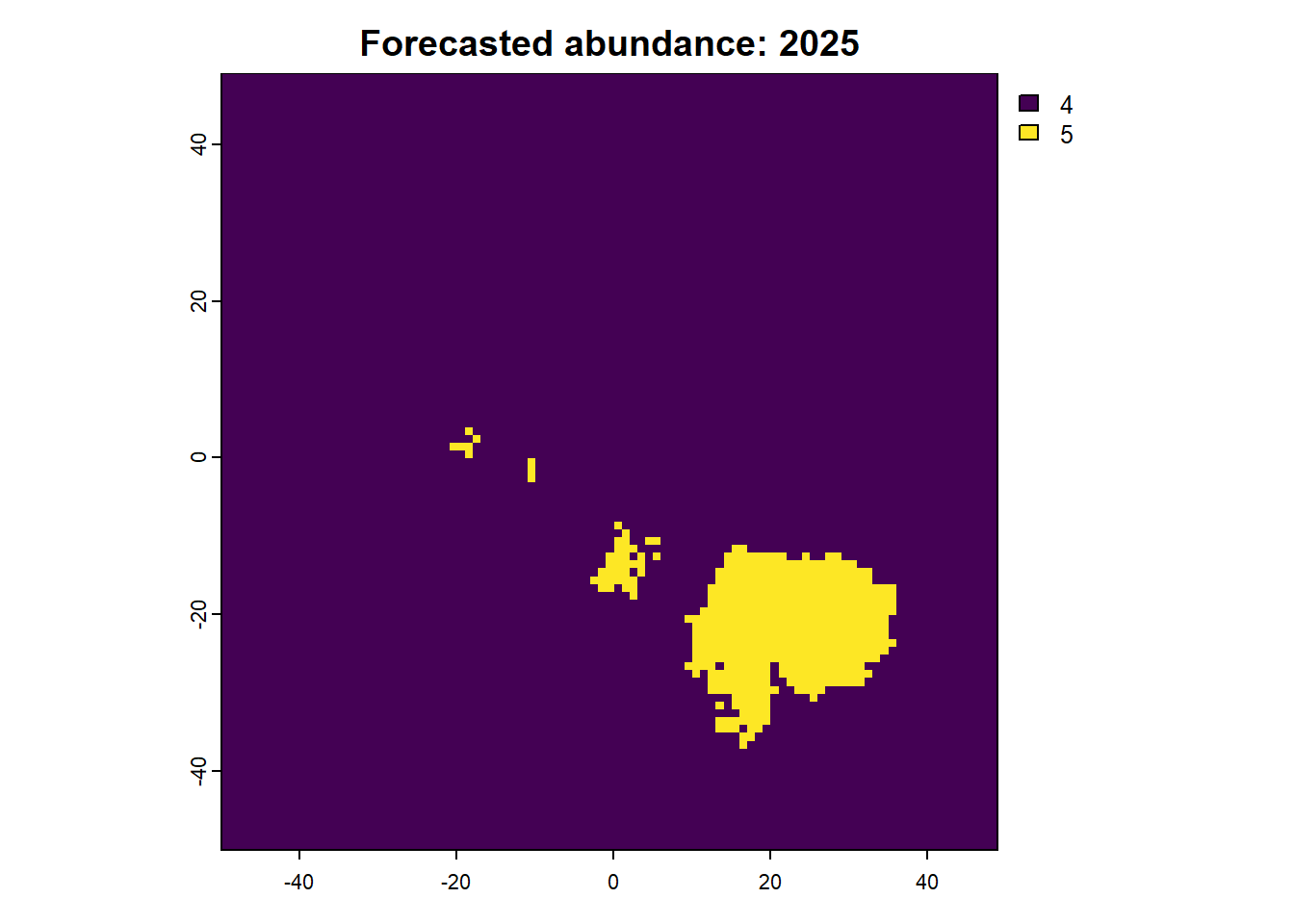
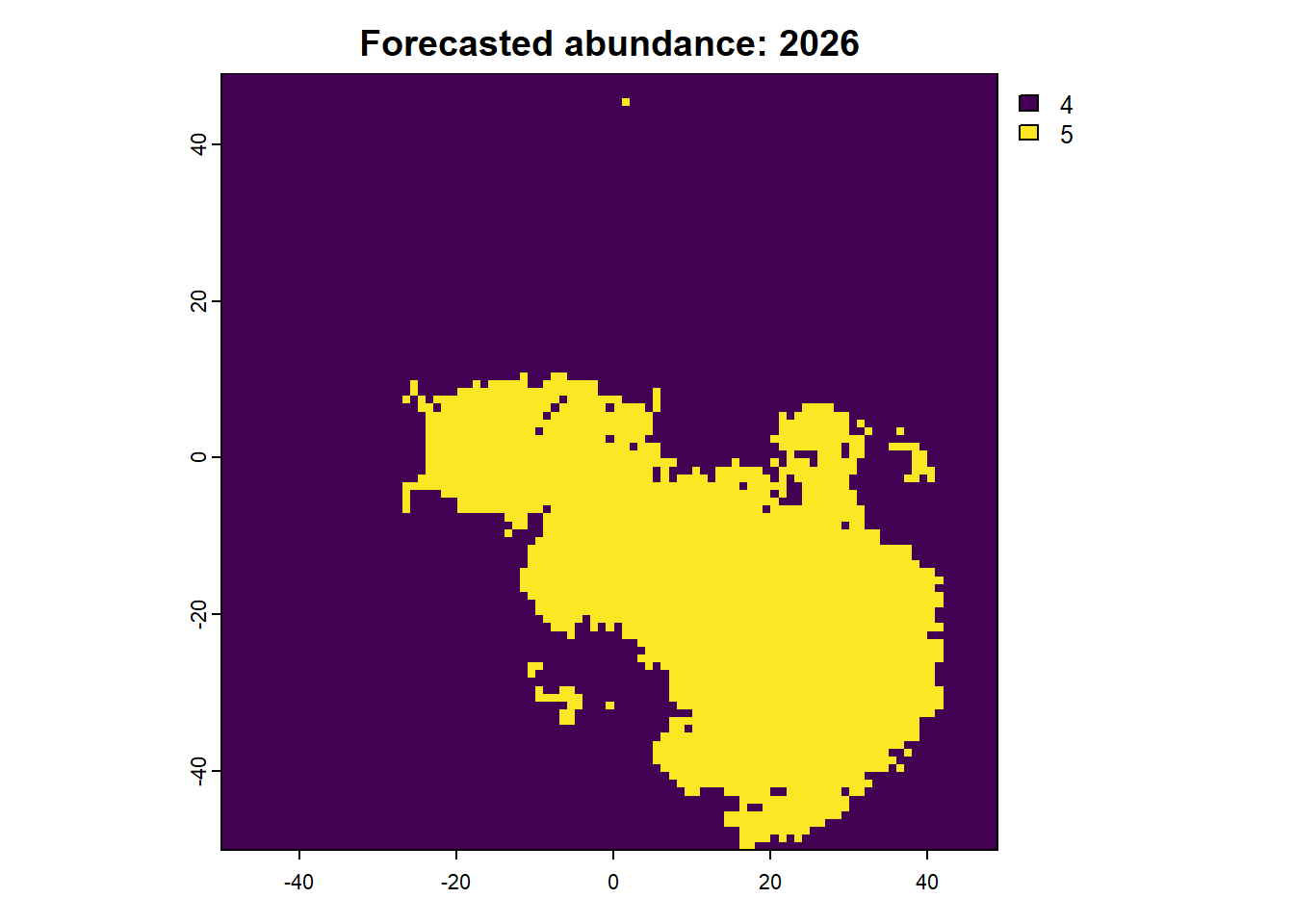
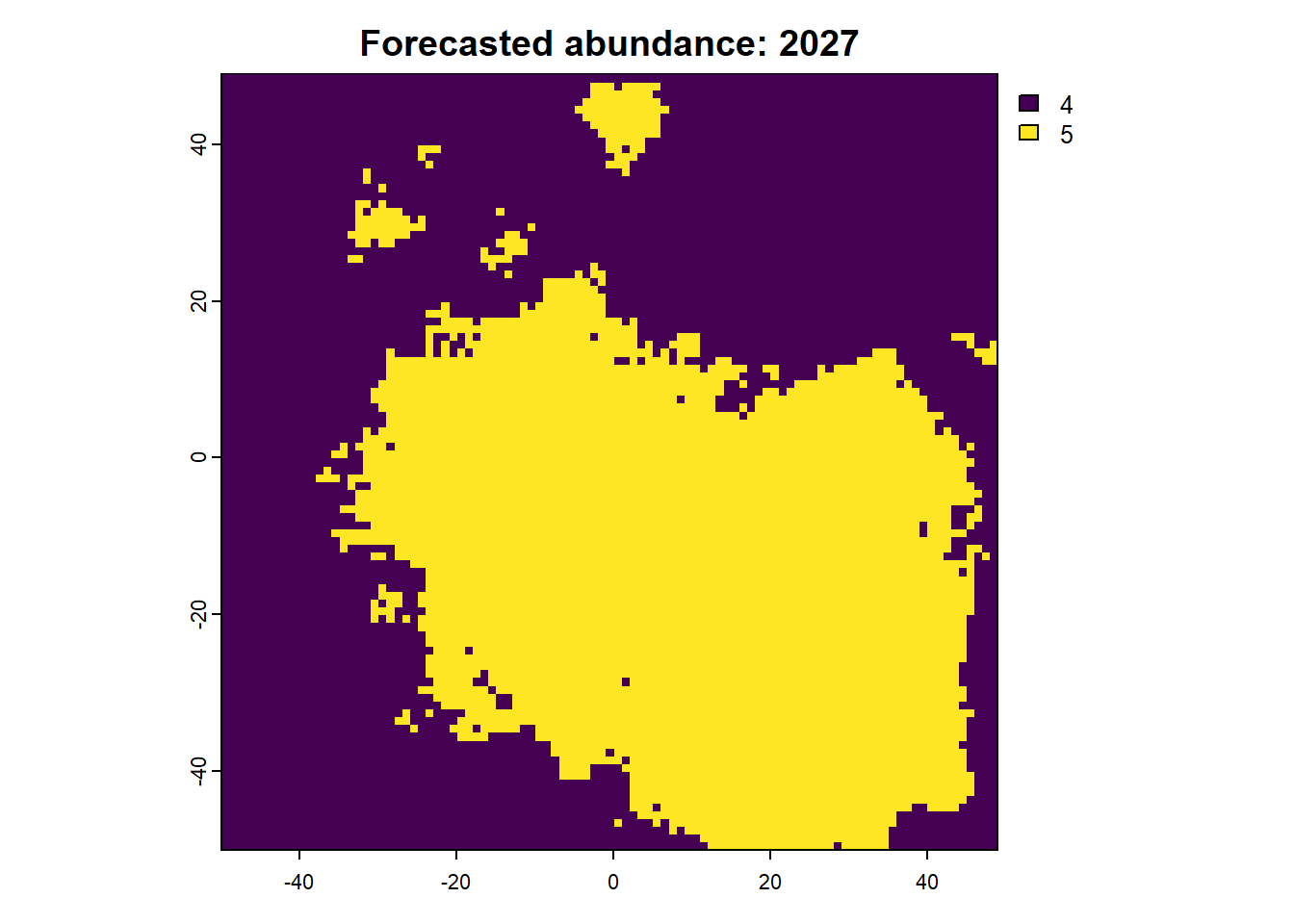
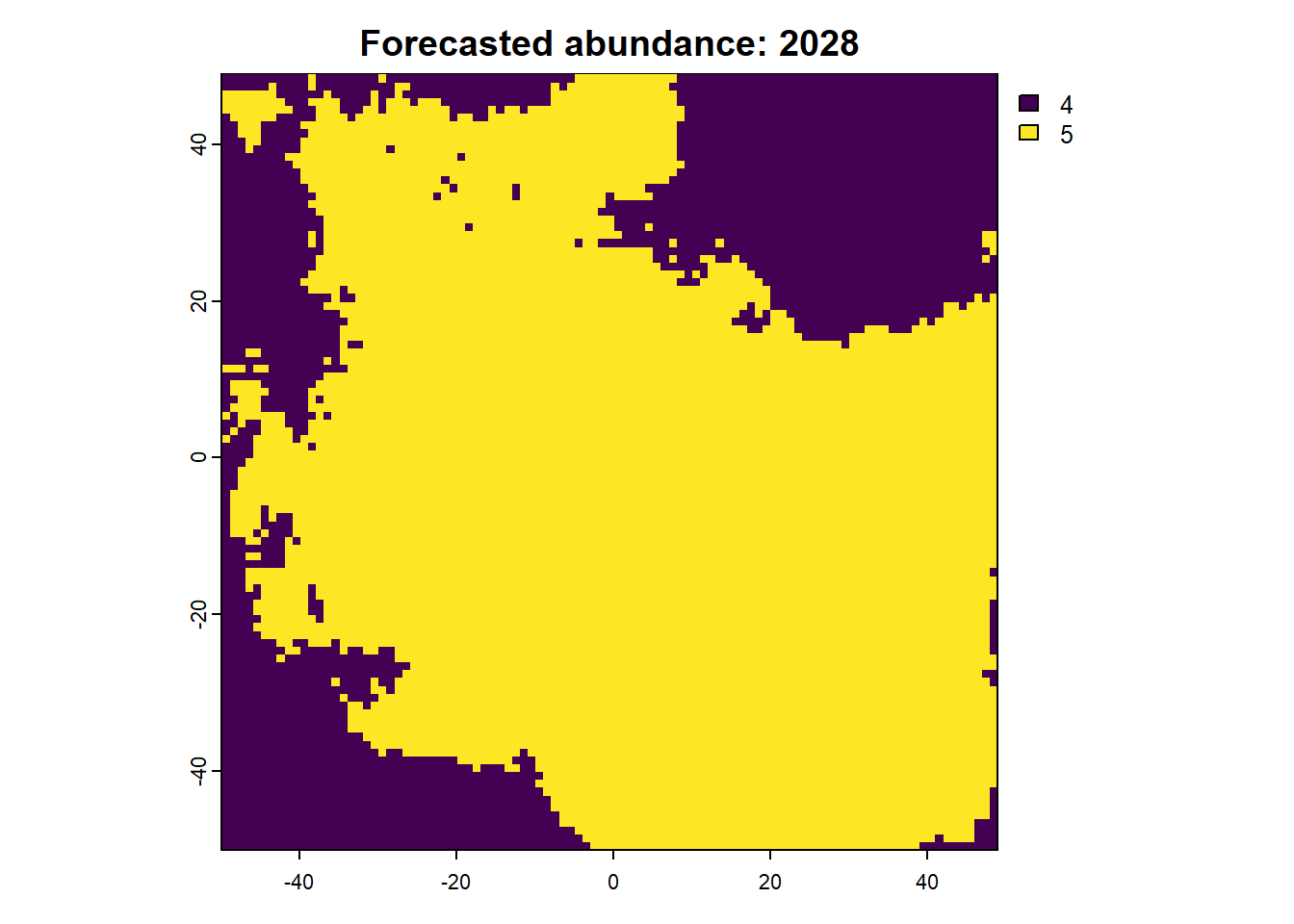
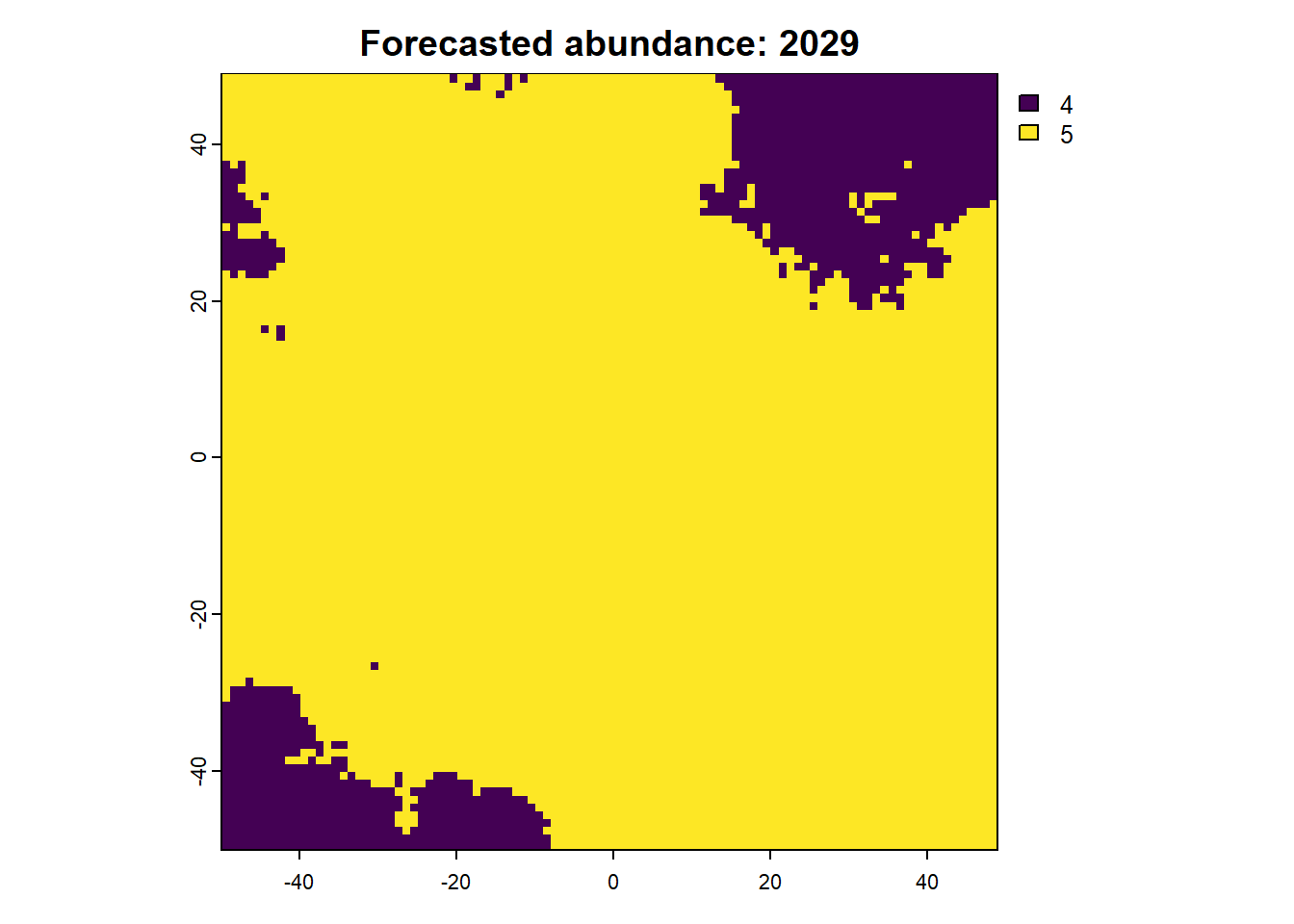
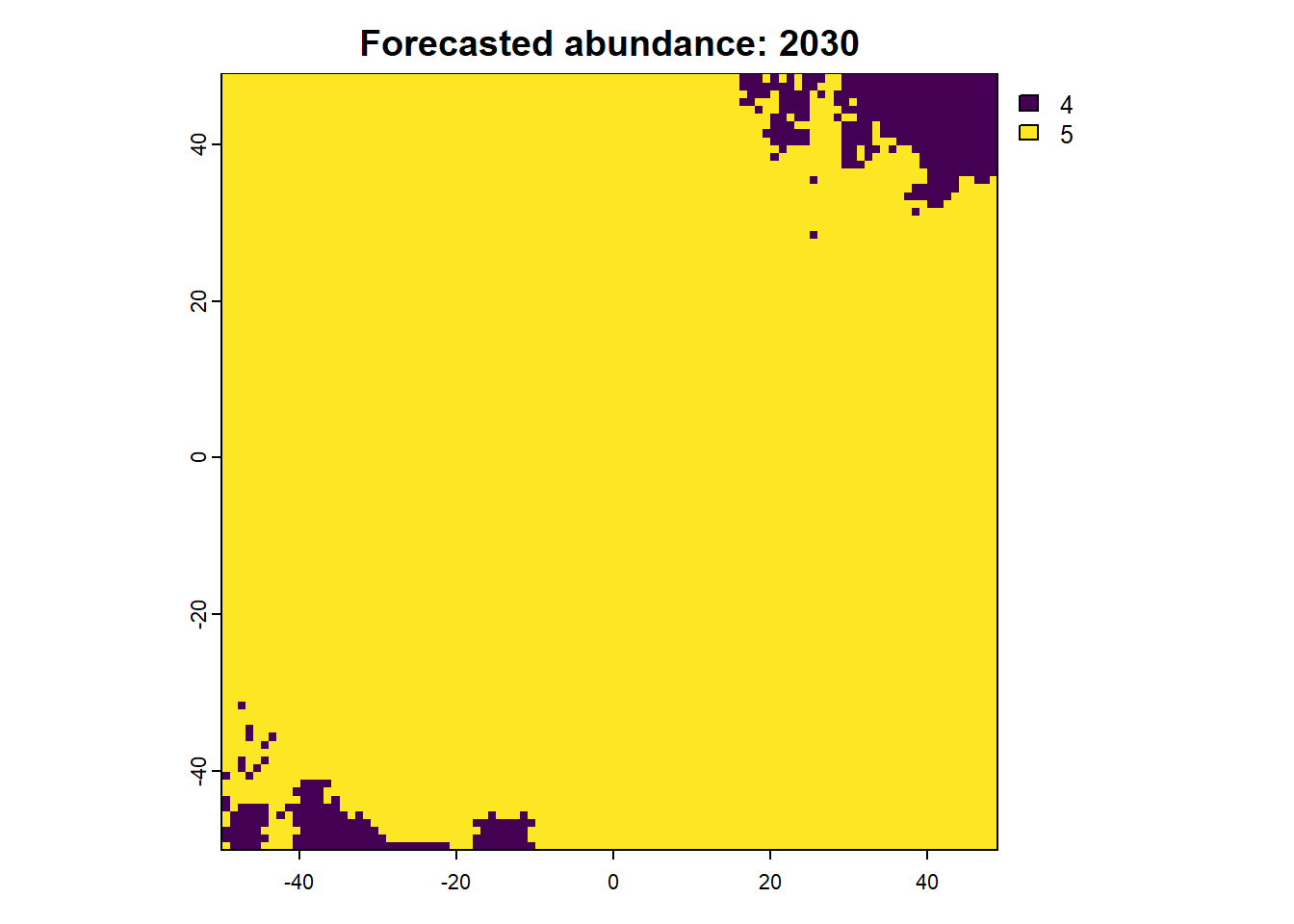
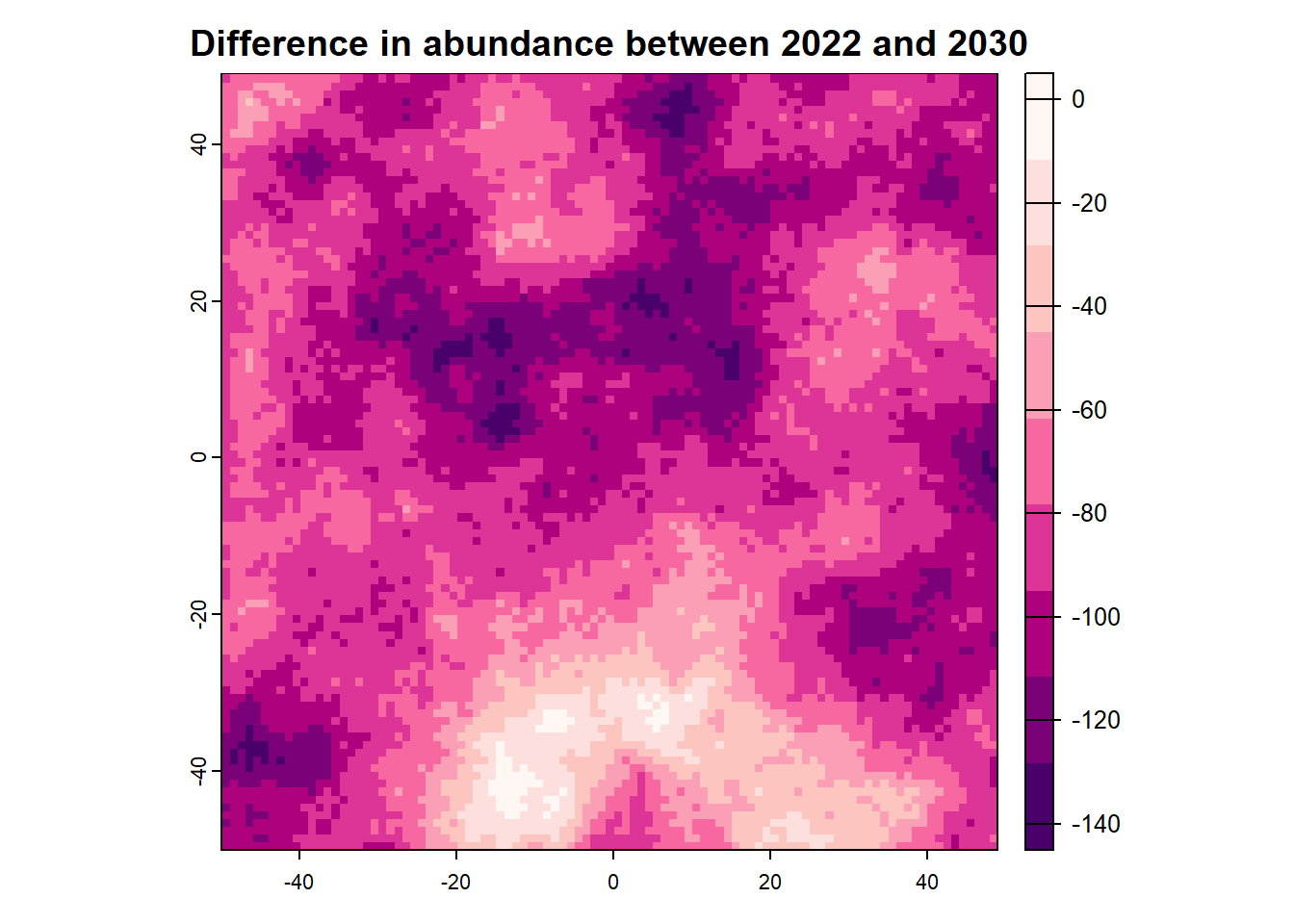
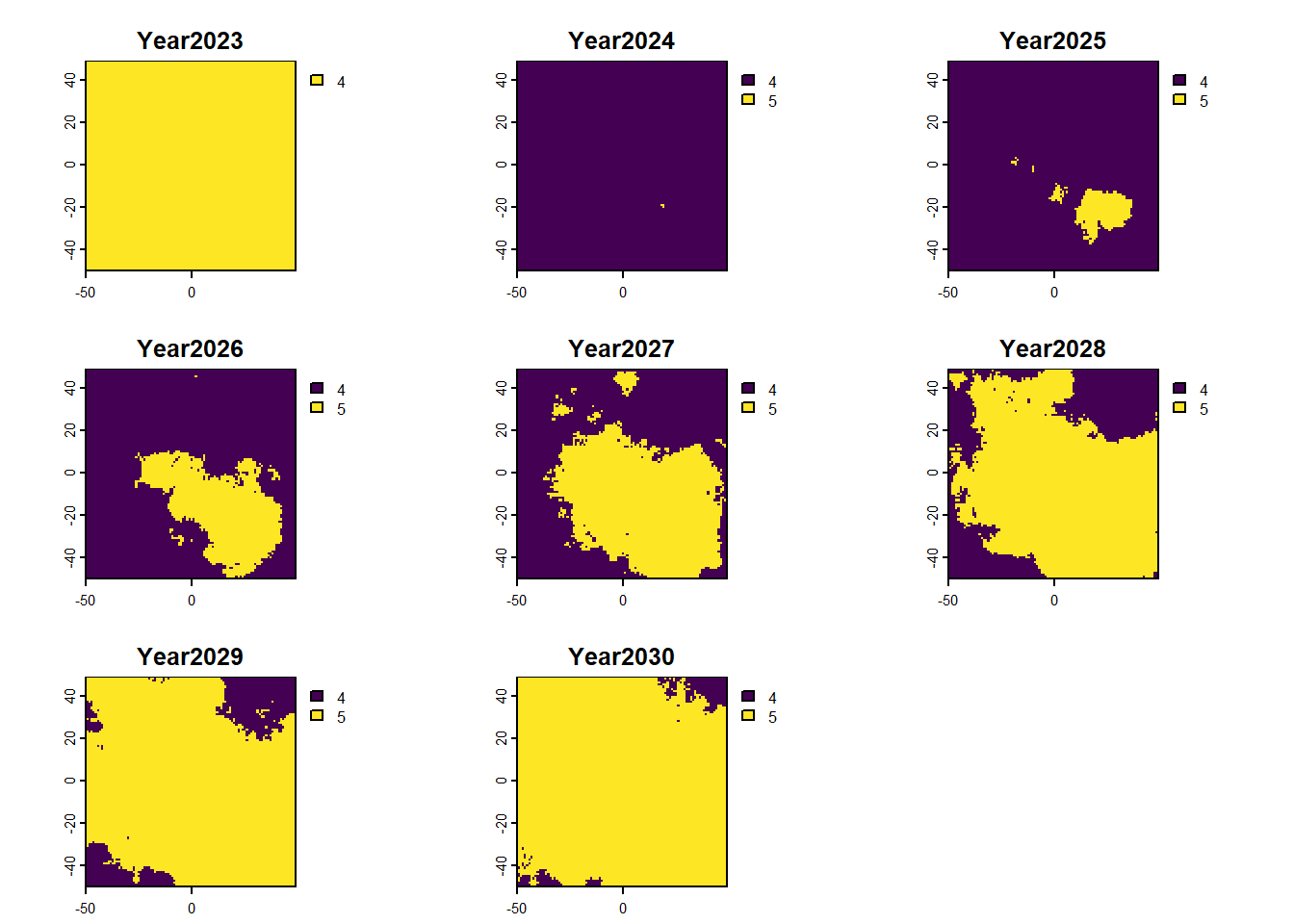
We can also access any objects and use them. For example, we can look at the forecasted abundances and the difference raster is also possible. This is done by calling the object name from the results list.
Code
:: plot (rast (results$ forecasts))
Examples
In the remaining chapters of this section, we will explore several examples, including learning of several arguments to the function setupProject. But we will start very simple first.
Barebones R script
Code
<- c ("predictiveecology.r-universe.dev" , getOption ("repos" ))# if (!require("SpaDES.project")) :: Install (c ("SpaDES.project" , "SpaDES.core" , "reproducible" ), repos = repos, dependencies = TRUE )= SpaDES.project:: .libPathDefault ("~/SpaDES_book/integratingSpaDESmodules" )= SpaDES.project:: .libPathDefault ("PredictiveEcology.org" )##################### PART II: Download the modules and install the needed packages <- SpaDES.project:: setupProject (paths = list (projectPath = "~/SpaDES_book/integratingSpaDESmodules" ,packagePath = packagePath),modules = c ("tati-micheletti/speciesAbundance@main" ,"tati-micheletti/temperature@main" ,"tati-micheletti/speciesAbundTempLM@main" ),times = list (start = 2013 ,end = 2030 ),updateRprofile = FALSE <- SpaDES.core:: simInitAndSpades2 (Setup):: completed (results):: moduleDiagram (results):: plot (rast (results$ forecasts))