# This repository is ours and it has the latest versions of our packages
repos <- c("predictiveecology.r-universe.dev", getOption("repos"))
# Need the latest version
if (tryCatch(packageVersion("SpaDES.project") < "0.1.1.9009", error = function(x) TRUE))
install.packages(c("SpaDES.project", "Require"), repos = repos)14 Forest Landscape Modelling with LandR and setupProject
See Barebones R script for the code shown in this chapter
2 hours – estimated time for 1st time executing (includes downloads)
24Gb RAM – estimated peak RAM for executing
93Gb disk – estimated hard drive allocation
LandR is a forest landscape model implemented as a collection of SpaDES modules in R. It is a reimplementation of LANDIS-II Biomass Succession Extension v.3.2.1, which at its core is very similar to v7. See the LandR Manual, Barros et al. (2023) and Scheller and Miranda (2015) for full details about forest dynamics simulated in LandR.
LandR fully open-source and users are expected to use, modify it and expand it (e.g. by creating new modules) as they see fit, as long has modifications are adequately reported. We hope that new modules are shared with others in the LandR community of users so that all can benefit.
Each LandR module is hosted in its own GitHub repository. When using and developing LandR modules, note that modules should be semi-independent. This is, they should be able to run error-free on their own, even they don’t produce useful outputs in this way. A good example are the “data preparation” and “data calibration” modules Biomass_speciesData and Biomass_borealDataPrep which can run on their own but will not produce any forest landscape simulations, only the input objects and parameters that Biomass_core (the “simulation module”) needs.
In this example, we will setup the workflow published in Barros et al. (2023) using setupProject from the SpaDES.project package and current versions of the LandR modules.
You need to have a Google account to access some of the data using the googledrive R package (part of the tidyverse family)1.
During the simInit() (or simInitAndSpades()) call R will prompt you to either choose a previously authenticated account (if you have previously used googledrive) or to open a browser window and authenticate.
Make sure you give tidyverse read/write access to your files:
We will eventually transition to not hosting data on Google. Until then, our apologies for any inconvenience.
14.1 All the steps of an ecological modelling project in a continuous workflow
14.2 Workflow setup
Show code
library(SpaDES.project)
out <- setupProject(
# OBJECTS needed within this function -----------------------------------
# these need to come *before* any formal arguments
# specifically these are needed for params.R
sppEquivCol = "Boreal",
vegLeadingProportion = 0,
# Named arguments for setupProject -- see ?SpaDES.project::setupProject for help
paths = list("packagePath" = file.path("packages", Require:::versionMajorMinor()),
"projectPath" = "~/SpaDES_book/LandRDemo_coreVeg"),
modules = c(
"PredictiveEcology/Biomass_speciesData@main"
, "PredictiveEcology/Biomass_borealDataPrep@main"
, "PredictiveEcology/Biomass_speciesParameters@main"
, "PredictiveEcology/Biomass_core@main"
# , "PredictiveEcology/Biomass_regeneration@main"
),
# packages only need to be specified if they are needed within this setupProject
# DiagrammeR is for plotting later in this qmd
packages = c("DiagrammeR", "googledrive", "httr2", "terra"),
options = list(
"reproducible.destinationPath" = paths$inputPath,
"spades.moduleCodeChecks" = FALSE,
"Require.cloneFrom" = Sys.getenv("R_LIBS_USER"), # faster package installs (from personal library)
"repos" = unique(repos)
),
# DYNAMIC MODEL SETUP ------------------------------------
times = list(start = 2001, end = 2031),
#
# # Module optional parameters
#params = "PredictiveEcology/PredictiveEcology.org@training-book/tutos/LandRDemo_coreVeg/params.R",
params = list(
.globals = list(sppEquivCol = "Boreal",
PSPdataTypes = "dummy")),
#
# # (more) INPUT OBJECTS -----------------------------------
# # these come after, so that we don't need to pre-install/load LandR
# # species lists/traits
studyArea = {
# create and use a random study area
# Lambert Conformal Conic for Canada: this is used in NRCan's "KNN" products
Biomass_corecrs <- "+proj=lcc +lat_1=49 +lat_2=77 +lat_0=0 +lon_0=-95 +x_0=0 +y_0=0 +datum=NAD83 +units=m +no_defs +ellps=GRS80 +towgs84=0,0,0"
centre <- terra::vect(cbind(-104.757, 55.68663), crs = "epsg:4326") # Lat Long
centre <- terra::project(centre, Biomass_corecrs)
studyArea <- LandR::randomStudyArea(centre, size = 2e8, seed = 1234)
},
# create a "larger" study area that can be used for parameter estimation, using
# more datasets that will fall into this larger area
studyAreaLarge = terra::buffer(studyArea, width = 3e4),
# sppEquiv is a table that defines equivalent names for species
# e.g., Pice_mar and Pice_Mar are identical
sppEquiv = {
speciesInStudy <- LandR::speciesInStudyArea(studyAreaLarge)
species <- LandR::equivalentName(speciesInStudy$speciesList,
df = LandR::sppEquivalencies_CA, sppEquivCol)
LandR::sppEquivalencies_CA[Boreal %in% species]
},
speciesParams = {
list(
"shadetolerance" = list(Lari_Lar = 1,
Pice_Gla = 2, Pice_Mar = 3,
Pinu_Ban = 1.5, Popu_Tre = 1
)
)
},
# OUTPUTS TO SAVE -----------------------
outputs = {
# save to disk 2 objects, every year
expand.grid(objectName = c("cohortData", "pixelGroupMap"),
saveTime = seq(times$start, times$end))
}
)If you have a look at other chapters in this section about setupProject, you will see some variation in the way we setup the workflows:
paths. Here we left the defaults for all paths (see?setupPaths()for the list of path options) except for the project location (projectPath) and the location of the package library (packagePath, which will be placed insideprojectPath).options. We also set a couple of “global options” that determine the where data will be downloaded to (“reproducible.destinationPath”). This will be the same as the default directory to look for inputs (“spades.inputPath”). Notice how we usedpaths$to get these directory paths from thepathsobject thatsetupProjectcreates (based on thepathsargument above) prior to setting the options.other arguments (
...). Almost all other arguments in the call above were part of the...(in thesetupProjectfunction). These are objects that are objects that are used by modules. Because modules have defaults for most objects, these tend to be “optionaly”; but the more a user understands a module, the more clearly the defaults can be evaluated as to whether they are sufficient or not. To avoid creating objects in the.GlobalEnvfirst, we take advantage ofsetupProject‘s ability to run the code in{ }and make these polygons. Note that these arguments are passed prior to any ’formal arguments’ (see?formalArgs())There are 4 objects that tend to be “required” in the modules that have emerged from the Predictive Ecology group. These are about the spatial area to be covered by the project: e.g.
studyArea,studyAreaLarge,rasterToMatchandrasterToMatchLarge. or whose defaults we want to override (e.g., the species table,sppEquiv, and trait values,speciesParams).
14.3 Run simulation
You can initialise the and run the workflow in two separate steps…
Code
# initialise then run simulation
# simInitOut <- SpaDES.core::simInit2(out)
simInitOut <- do.call(SpaDES.core::simInit, out)
simOut <- SpaDES.core::spades(simInitOut)… or together, using simInitAndSpades2()
Code
simOut <- SpaDES.core::simInitAndSpades2(out)14.3.1 Inspect the workflow, its inputs, outputs and parameters
Now that we have a simulation we can inspect it. Let’s assume you initialised and ran your simulation with two separate simInit2() and spades() calls.
Inspect module connections and between-module object flows:
Code
SpaDES.core::moduleDiagram(simInitOut)
SpaDES.core::objectDiagram(simInitOut) 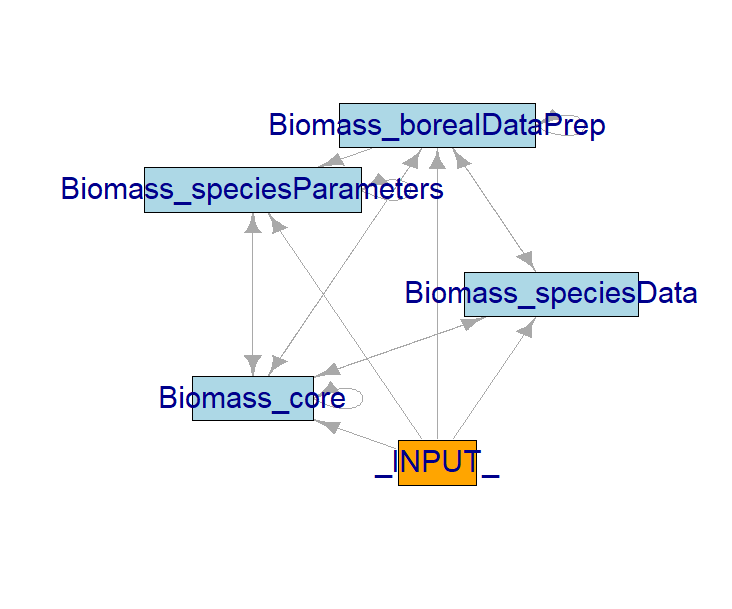
Note the scheduling of the init events in simInitOut and how simOut has future events scheduled too – thanks to this, we can extend the simulation beyond the original SpaDES.core::end(sim) of 2031 (Extend the simulation).
Code
SpaDES.core::events(simInitOut)
SpaDES.core::events(simOut)We can also have a look at the completed events:
Code
SpaDES.core::completed(simInitOut)
SpaDES.core::completed(simOut)Finally, the following accessor functions will show a list of model inputs, outputs and parameters:
Code
SpaDES.core::inputs(simOut)
SpaDES.core::outputs(simOut)
SpaDES.core::parameters(simOut)We could even plot some of the input and output rasters to check that they are as we expected – no need to look for these objects files, they are all in the simList.
Code
# spatial inputs from list above
terra::plot(simOut$studyAreaLarge, col = "navyblue", main = "studyArea & studyAreaLarge")
terra::plot(simOut$studyArea, col = "lightblue", add = TRUE)
# spatial outputs from list above
terra::plot(simOut$vegTypeMap,
col = hcl.colors(palette = "Dynamic", n = length(unique(simOut$vegTypeMap[]))),
main = "", add = TRUE)
terra::plot(simOut$speciesLayers)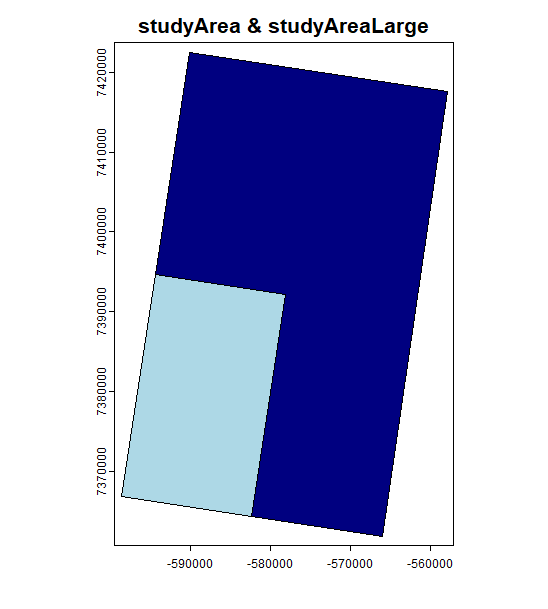
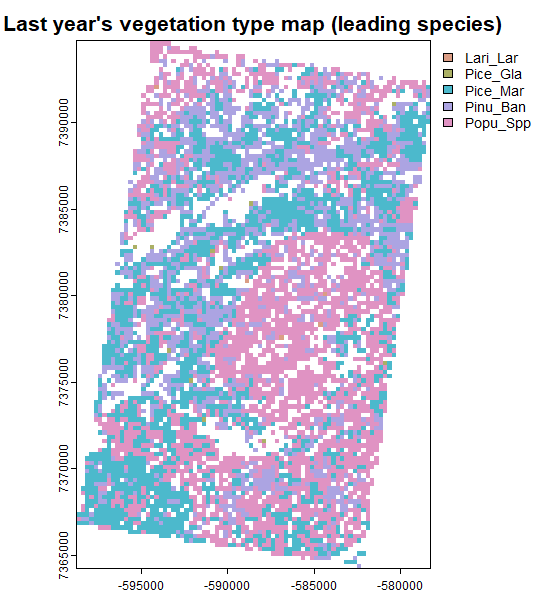
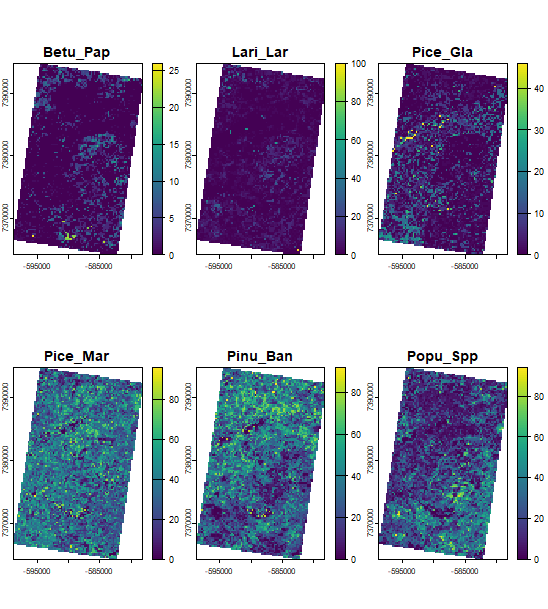
Inspecting inputs and outputs directly from the simList
More importantly in our view, is the ability to inspect statistical models used to fit model parameters. This is possible because the developers have declared the fitted statistical model objects as module outputs. Often, this type of information is buried in supplementary materials of papers and incomplete (e.g. coefficients and goodness-of-fit statistics are presented, but the entire model object, with its fitted values, residuals, etc., are not).
By exporting entire model objects, and making them available via repeatable code or data repositories, model transparency and potential scrutiny are massively increased.
Code
# model used to estimate species establishment probabilities
summary(simOut$modelBiomass$mod)
plot(simOut$modelBiomass$mod)
# model used to calibrate Picea glauca's growth parameters
summary(simOut$speciesGrowthCurves$Pice_Gla$NonLinearModel$Pice_Gla)14.3.2 Turn plotting on after setting up the workflow
We can change parameters and re-run the simulation to, e.g., activate live plotting in Biomass_core, without having to
change the parameter provided to
setupProjectrepeat the
setupProjectcall
Note that because simOut has some objects that have “pointers”, like SpatRaster objects that come from tif files, we may not be able to start from the previous simInitOut. In other words, even though it looks like it is the object that emerges from the simInit function and therefore, we should be able to pass it to spades, it may not work as desired (can test this, if desired). This means that simInitOut may need to re-generated, otherwise spades would inherit some objects that were changed during the previous spades.
Thanks to internal caching, it will only take seconds to “redo” simInitOut. You will also notice that init events are retrieved from cache, this time around2.
Code
simInitOut <- SpaDES.core::simInit2(out)
SpaDES.core::P(simInitOut, param = ".plots", module = "Biomass_core") <- "screen"
SpaDES.core::P(simInitOut, param = ".plotMaps", module = "Biomass_core") <- TRUE
simOut <- SpaDES.core::spades(simInitOut)14.3.3 Extend the simulation
We can also keep it going for a few more years. Use SpaDES.core::end() to extend the simulation another 20 years and then call SpaDES.core::spades() on the changed simList (not the one output by `SpaDES.core::simInit2()) to resume the simulation from 2031.
Code
SpaDES.core::end(simOut) <- 2061
simOut <- SpaDES.core::spades(simOut)14.3.4 Many replicates (somewhat experimental; actively developing)
Dynamic ecological models like LandR often simulate stochastic ecological processes, like dispersal, probability of germination, or fire spread (see Chapter 15), which requires replicating simulations. The number of replicates will largely depend on the variability generated by the model. The more variability, the more replicates needed.
The set of LandR modules used here generates little variation in tree species dynamics and 10 replicates are sufficient.
We use SpaDES.experiment::experiment2() to do two things:
Create a folder structure by replicate to store the simulation outputs
Generate 2 independent simulations and run them in sequence (Listing 14.1) or in parallel (Listing 14.2), using the
futurepackage.
Require::Install("SpaDES.experiment (HEAD)", repos = unique(repos))
simInitOut <- SpaDES.core::simInit2(out)
future::plan("sequential")
simOutExperiment <- SpaDES.experiment::experiment2(simInitOut, replicates = 2)14.3.4.1 Parallel
This works, in theory, on Windows, Linux and MacOS, but in practice seems to be less problematic on Linux.
future::plan("multisession", workers = 2)
simOutExperiment <- SpaDES.experiment::experiment2(simInitOut, replicates = 2)
future:::ClusterRegistry("stop") In general, there are two levels of replicate independence that can be considered in dynamic ecological models like LandR, where stocahstic processes happen during the parameterisation phase[^3] and during the simulation phase:
replicates may have independent parameterisation and initialisation and independent ecological process simulation
replicates may share the parameterisation and initialisation but have independent ecological process simulation.
Neither is better than the other and the one to chose depends on the questions and hypotheses being asked.
In the example above, replicates share the same parameters/initial conditions because they all access the same cached objects from the init events – scroll up to see printed outputs from experiment2().
To have fully independent replication we can use experiment2(..., useCache = FALSE)
14.4 Validation (very experimental)
Now that we have several replicates, we can use Biomass_validationKNN to run some validation statistics.
Integrating model validation as part of an ecological modelling workflow ensures that when model predictions are repeated under the same or different conditions, or new data arrives for validation, they can be readily (re-evaluated).
Note that, for convenience, we use simOut to access many of the necessary inputs for Biomass_validationKNN, but we could have used one of the simLists in simOutExperiment instead.
We pass all raster objects from the saved files directly, to ensure they are linked to these file paths explicitly.
Code
outValid <- setupProject(
paths = list("packagePath" = "packages/",
"projectPath" = "~/SpaDES_book/LandRDemo_coreVeg",
"outputPath" = "validation/"),
options = list(
"LandR.assertions" = TRUE,
"reproducible.destinationPath" = paths$inputPath,
"spades.inputPath" = paths$inputPath,
"spades.moduleCodeChecks" = FALSE,
"repos" = repos
),
modules = c(
"PredictiveEcology/Biomass_validationKNN@main"
),
# SIMULATION SETUP --------------------------------
times = list(start = 1, end = 1),
# PARAMETERS --------------------------------------
params = list("Biomass_validationKNN" = list(
"minCoverThreshold" = SpaDES.core::params(simOut)$Biomass_borealDataPrep$minCoverThreshold,
"pixelGroupBiomassClass" = SpaDES.core::params(simOut)$Biomass_borealDataPrep$pixelGroupBiomassClass,
"deciduousCoverDiscount" = SpaDES.core::params(simOut)$Biomass_borealDataPrep$deciduousCoverDiscount,
"sppEquivCol" = SpaDES.core::params(simOut)$Biomass_borealDataPrep$sppEquivCol,
".plots" = c("png") # save all to .png
)),
# INPUT OBJECTS -----------------------------------
studyArea = {
simOut$studyArea
},
rasterToMatch = {
terra::deepcopy(simOut$rasterToMatch)
},
rawBiomassMapStart = {
terra::deepcopy(simOut$rasterToMatchLarge)
},
simulationOutputs = {
lapply(simOutExperiment, SpaDES.core::outputs) |>
rbindlist()
},
sppEquiv = simOut$sppEquiv,
sppEquivCol = simOut$sppEquivCol,
sppColorVect = simOut$sppColorVect,
speciesLayersStart = {
terra::deepcopy(simOut$speciesLayers)
},
standAgeMapStart = {
terra::deepcopy(simOut$standAgeMap)
}
)
simOutValid <- SpaDES.core::simInitAndSpades2(outValid)14.5 Different scenarios and model selection
In Barros et al. (2023), the model was run with two different parameterisation approaches one that was “data hungry” and calibrated tree species growth parameters (using Biomass_speciesParameters as we have done above) and a simpler one that used default parameter values (without Biomass_speciesParameters).
SpaDES and LandR allow us to swap parameterisation/calibration approaches easily and re-evaluate each and compare the models (see Barros et al. (2023)).
Do to this, we simply exclude Biomass_speciesParameters from out and run a second simulation. We also need to save the outputs in a different folder, or the previous ones will be overriden.
Code
out2 <- out
out2$modules <- out2$modules[out2$modules != "Biomass_speciesParameters"]
out2$paths$outputPath <- normPath(file.path("~/SpaDES_book/LandRDemo_coreVeg", "outputsSim2"))
simOut2 <- SpaDES.core::simInitAndSpades2(out2)14.6 Debugging
There are several ways to debug SpaDES modules (see Chapter 12), a relatively easy one for when you are suprised by an error occurring during specific event is to pass the event’s name to spades(..., debug = ) argument.
Below, we debug the plotSummaryBySpecies event of Biomass_core. R interrupts the execution of the code in the chunk that executes this event’s operations (inside doEvent.Biomass_core())
Code
simOut <- SpaDES.core::spades(simInitOut, debug = "plotSummaryBySpecies")R interrupts code execution during this event’s operations (inside doEvent.Biomass_core()):
debugging in: get(moduleCall, envir = fnEnv)(sim, cur[["eventTime"]], cur[["eventType"]])
debug at C:/Users/cbarros/Documents/SpaDES_book/LandRDemo_coreVeg/modules/Biomass_core/Biomass_core.R#314: {
(...)From there you can press ENTER, F10 or the “Next” button to execute the code line-by-line. At some point you will get to this line:
Code
sim <- plotSummaryBySpecies(sim)which calls the function that effectively makes the summary plots. If you spotted a problem during the plotSummaryBySpecies event (or maybe you want to see what it does and/or change the code) and it hasn’t been triggered yet, then it’s likely it happened in this function.
Before running the line you can debugonce(plotSummaryBySpecies) to enable debugging the function and spot the issue.
Another option would be to insert a browser() at the top of the function’s definition inside the module code or the R scripts in the module’s R/ folder (<modulePath>/<module_name>/R/). In this case look for plotSummaryBySpecies <- compiler::cmpfun(function(sim) {...} inside the module code (Biomass_core.R) and try putting a browser() inside the {}
14.7 Try on your own
Try re-running the workflow with a different set of study areas. For example:
Code
# studyArea could be
studyArea = {
set.seed(123)
SpaDES.tools::randomStudyArea(size = 200000000)
}
# studyAreaLarge
studyAreaLarge = {
terra::buffer(studyArea, width = 10000)
}Noticed any differences (speed, cache IDs, …)?
were the species simulated the same? How about their trait values (e.g. estimated maxB, species establishment probabilities. )
14.8 See also
Chapter 10 on caching.
14.9 Barebones R script
Code
library(SpaDES.project)
out <- setupProject(
# OBJECTS needed within this function -----------------------------------
# these need to come *before* any formal arguments
# specifically these are needed for params.R
sppEquivCol = "Boreal",
vegLeadingProportion = 0,
# Named arguments for setupProject -- see ?SpaDES.project::setupProject for help
paths = list("packagePath" = file.path("packages", Require:::versionMajorMinor()),
"projectPath" = "~/SpaDES_book/LandRDemo_coreVeg"),
modules = c(
"PredictiveEcology/Biomass_speciesData@main"
, "PredictiveEcology/Biomass_borealDataPrep@main"
, "PredictiveEcology/Biomass_speciesParameters@main"
, "PredictiveEcology/Biomass_core@main"
# , "PredictiveEcology/Biomass_regeneration@main"
),
# packages only need to be specified if they are needed within this setupProject
# DiagrammeR is for plotting later in this qmd
packages = c("DiagrammeR", "googledrive", "httr2", "terra"),
options = list(
"reproducible.destinationPath" = paths$inputPath,
"spades.moduleCodeChecks" = FALSE,
"Require.cloneFrom" = Sys.getenv("R_LIBS_USER"), # faster package installs (from personal library)
"repos" = unique(repos)
),
# DYNAMIC MODEL SETUP ------------------------------------
times = list(start = 2001, end = 2031),
#
# # Module optional parameters
#params = "PredictiveEcology/PredictiveEcology.org@training-book/tutos/LandRDemo_coreVeg/params.R",
params = list(
.globals = list(sppEquivCol = "Boreal",
PSPdataTypes = "dummy")),
#
# # (more) INPUT OBJECTS -----------------------------------
# # these come after, so that we don't need to pre-install/load LandR
# # species lists/traits
studyArea = {
# create and use a random study area
# Lambert Conformal Conic for Canada: this is used in NRCan's "KNN" products
Biomass_corecrs <- "+proj=lcc +lat_1=49 +lat_2=77 +lat_0=0 +lon_0=-95 +x_0=0 +y_0=0 +datum=NAD83 +units=m +no_defs +ellps=GRS80 +towgs84=0,0,0"
centre <- terra::vect(cbind(-104.757, 55.68663), crs = "epsg:4326") # Lat Long
centre <- terra::project(centre, Biomass_corecrs)
studyArea <- LandR::randomStudyArea(centre, size = 2e8, seed = 1234)
},
# create a "larger" study area that can be used for parameter estimation, using
# more datasets that will fall into this larger area
studyAreaLarge = terra::buffer(studyArea, width = 3e4),
# sppEquiv is a table that defines equivalent names for species
# e.g., Pice_mar and Pice_Mar are identical
sppEquiv = {
speciesInStudy <- LandR::speciesInStudyArea(studyAreaLarge)
species <- LandR::equivalentName(speciesInStudy$speciesList,
df = LandR::sppEquivalencies_CA, sppEquivCol)
LandR::sppEquivalencies_CA[Boreal %in% species]
},
speciesParams = {
list(
"shadetolerance" = list(Lari_Lar = 1,
Pice_Gla = 2, Pice_Mar = 3,
Pinu_Ban = 1.5, Popu_Tre = 1
)
)
},
# OUTPUTS TO SAVE -----------------------
outputs = {
# save to disk 2 objects, every year
expand.grid(objectName = c("cohortData", "pixelGroupMap"),
saveTime = seq(times$start, times$end))
}
)
# initialise then run simulation
# simInitOut <- SpaDES.core::simInit2(out)
simInitOut <- do.call(SpaDES.core::simInit, out)
simOut <- SpaDES.core::spades(simInitOut)
simOut <- SpaDES.core::simInitAndSpades2(out)
SpaDES.core::moduleDiagram(simInitOut)
SpaDES.core::objectDiagram(simInitOut)
SpaDES.core::events(simInitOut)
SpaDES.core::events(simOut)
SpaDES.core::completed(simInitOut)
SpaDES.core::completed(simOut)
SpaDES.core::inputs(simOut)
SpaDES.core::outputs(simOut)
SpaDES.core::parameters(simOut)
# spatial inputs from list above
terra::plot(simOut$studyAreaLarge, col = "navyblue", main = "studyArea & studyAreaLarge")
terra::plot(simOut$studyArea, col = "lightblue", add = TRUE)
# spatial outputs from list above
terra::plot(simOut$vegTypeMap,
col = hcl.colors(palette = "Dynamic", n = length(unique(simOut$vegTypeMap[]))),
main = "", add = TRUE)
terra::plot(simOut$speciesLayers)
# model used to estimate species establishment probabilities
summary(simOut$modelBiomass$mod)
plot(simOut$modelBiomass$mod)
# model used to calibrate Picea glauca's growth parameters
summary(simOut$speciesGrowthCurves$Pice_Gla$NonLinearModel$Pice_Gla)
simInitOut <- SpaDES.core::simInit2(out)
SpaDES.core::P(simInitOut, param = ".plots", module = "Biomass_core") <- "screen"
SpaDES.core::P(simInitOut, param = ".plotMaps", module = "Biomass_core") <- TRUE
simOut <- SpaDES.core::spades(simInitOut)
SpaDES.core::end(simOut) <- 2061
simOut <- SpaDES.core::spades(simOut)
out2 <- out
out2$modules <- out2$modules[out2$modules != "Biomass_speciesParameters"]
out2$paths$outputPath <- normPath(file.path("~/SpaDES_book/LandRDemo_coreVeg", "outputsSim2"))
simOut2 <- SpaDES.core::simInitAndSpades2(out2)
simOut <- SpaDES.core::spades(simInitOut, debug = "plotSummaryBySpecies")
sim <- plotSummaryBySpecies(sim)
# studyArea could be
studyArea = {
set.seed(123)
SpaDES.tools::randomStudyArea(size = 200000000)
}
# studyAreaLarge
studyAreaLarge = {
terra::buffer(studyArea, width = 10000)
}See
?googledrive::drive_authfor more information.↩︎See Chapter 10 about caching.↩︎