Introduction
This book presents a collection of approaches and examples that aim to facilitate the building of nimble workflows for science-based problems that follow in the principles of repeatability, reproducibility, reusability and transparency (R3T). We present these using the SpaDES family of packages.
Why SpaDES?
SpaDES is a set of R packages that facilitate implementing nimble workflows that follow in the principles of repeatability, reproducibility, reusability and transparency (R3T), via the creation and use of highly modular code that has metadata attached.
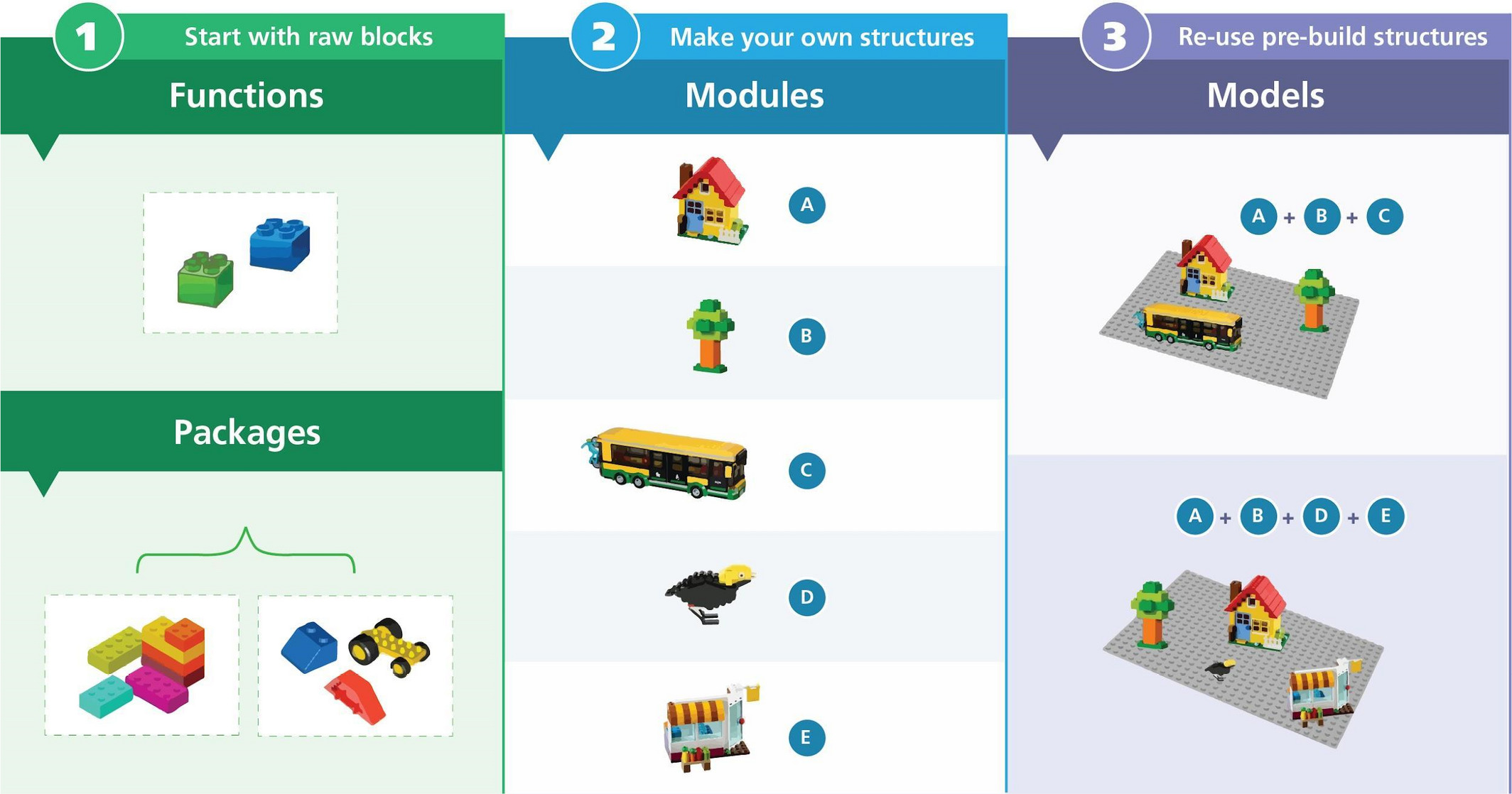
Modularity
Modular code for ecological analyses has been long proposed (Reynolds and Acock 1997). Modularity means that scripts can be broken up into themes that bundled into meaningful chunks that can be easily moved, swapped, replaced or scrapped easily. A nice example of this is Barros et al. (2023) where a module was included or removed to identify the value of “adding more complexity”. One simple conclusion followed Occam’s Razor: simpler data and models predicted simpler characteristics of the forest more accurately than the complex data and models. In other words, adding complexity to a model made the model perform worse for some characteristics.
Metadata
Metadata in a SpaDES context includes identifying what objects are expected by a chunk of code and what objects are created by a chunk of code: the inputs and outputs. This forces a user to understand their code and its linkages, and it allows R to know how to connect this chunk of code with others. Importantly, it allows SpaDES to chain modules such that the workflow structure can be defined from the bottom-up, rather than top-down.
Nimbleness
Together SpaDES’s modularity, standardized structure and metatada, improve workflow nimbleness, where we can weave together new Results that use new data, updated data, a new module, a new study area, a new visualization, address new stakeholder’s needs and more.
Aren’t packages and functions sufficient?
Scientists have embraced the idea of modular code by creating and using functions, often bundled into packages, e.g., R packages. This is analogous to the Methods section of a journal paper: i.e., the methods (functions) describe how you will do something. The use of packages in this way has massively accelerated Ecology and other data sciences.
However, even if you know all the functions that a paper used, this in itself is not sufficient to reproduce the Results. SpaDES modules can be thought of the recipe needed to make the Results. In other words, a SpaDES module enables a developer to bundle the functions in a specific order so that one can recreate the steps to make the Results. A user can then use the module directly, rather instead of trying to recreate the necessary sequence of functions.
I use targets for my workflows. How does SpaDES differ?
targets is an R package (descendant of drake), very similar to GNU Make, that facilitates building data analyses pipelines.
There are two major differences between targets and the SpaDES approach:
targetsis “top-down”, SpaDES is “bottom-up” – withtargets, the workflow is entirely defined by the script developer (as in Make). That is, they define the connections and the sequence between each “target” (the code sections that call functions and execute operations), when they will be iterated and for “how many times”. Re-arranging “targets” sequences and iterations may therefore be hard, if the “target” code is not well known/understood.SpaDES is meant to enable mixing-and-matching different modules that share the same inputs/outputs easily – i.e. without having to have deep knowledge of the module code. SpaDES will read each module’s metadata and from it deduce the sequence in which modules need to be chained. SpaDES will also iterate module code when necessary, following the developer’s “scheduling” instructions. The workflow sequence, therefore, arises from the set of modules that is being used1.
SpaDES has “embedded” metadata – or at least it makes the developer think about it2. Each module is defined by listing not only the inputs and outputs, but also their descriptions and types of object class, the module’s description, its operating time scale, package and module dependencies, and much more. Some of the metadata is essential to chain modules (lists of inputs and outputs), some is not but can greatly facilitate workflow setup (e.g. declaring module dependencies), while other metadata is simply good to have (e.g. a description of the module).
Finally, both targets and SpaDES have embedded caching mechanisms, which in SpaDES can be controlled at several levels (by the user, inside the module code, etc.) – see 10 Introduction to Cache on SpaDES caching mechanisms.
SpaDES & applied ecology
SpaDES development was, and continues to be, strongly motivated by our attempts to answer important issues in applied ecology.
Alongside these problems, ecologists must grapple with new expectations for how they do their work to inform these problems.
New expectations
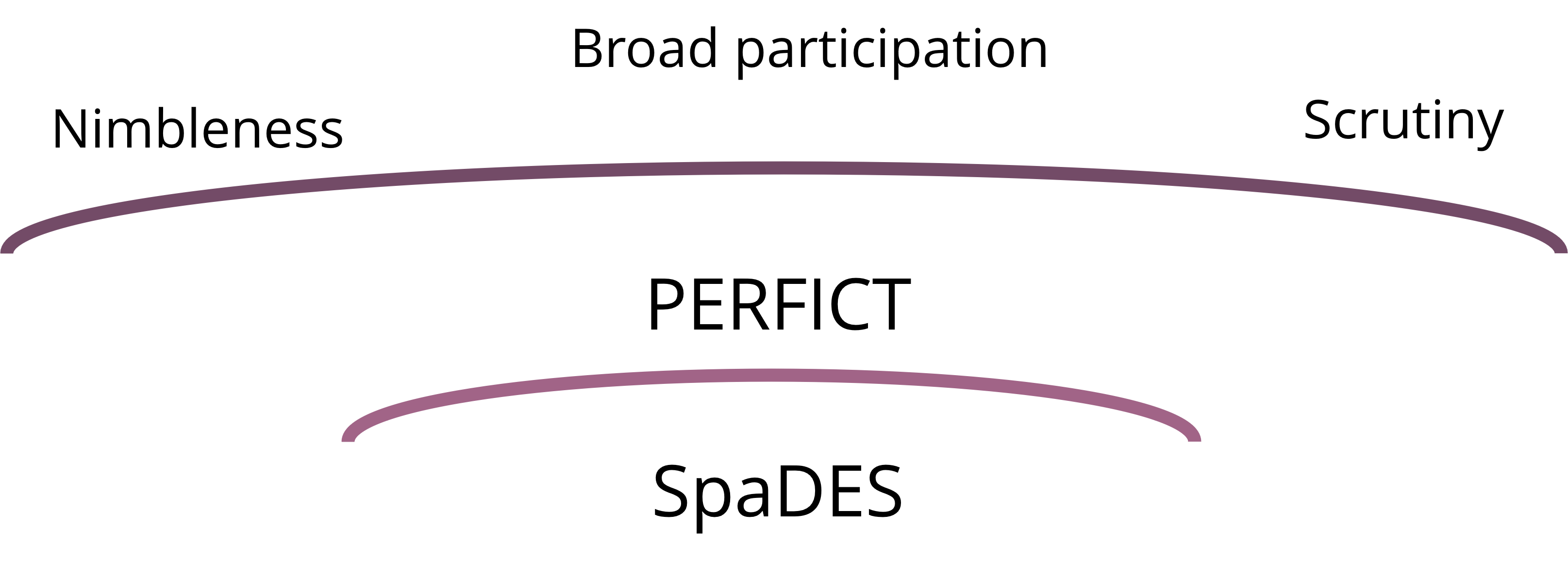
Applied ecologists of the 21st century are expected more and more to uphold standards of nimbleness, broad participation and scrutiny.
Analyses must be able to respond quickly to new data, new insights, new drivers, and new management needs; projects must include knowledge and participation from outside the scientific community and be relevant to non-scientific audiences; and they must accept and embrace scientific and non-scientific scrutiny.
These expectations are not independent of each other. Greater nimbleness allows for easier participation by others and scrutiny benefits from the participation of a broad range of specialists and non-specialists.
Meeting these expectations requires the R3T:
repeatability – analyses must be able to produce the same results, when using the exact same context (e.g. same user, same machine setup);
reproducibility – analyses must be able to produce the same results by others and into the future;
reusability – the methodology must be easy to transfer and expand in a different context;
transparency – analyses must be able to be inspected, understood, and scrutinized by many eyes;
but also
forecasting – analyses must demonstrate that they are proving reliable insights into the future, not just the past or theoretical expectation;
validation – analyses must clearly demonstrate why they should be trusted and used;
open-data/open-models – raw data, parameters, model algorithms have to be (freely) accessible as much as possible;
testing – models and predictions should be continuously tested.
Together these 10 requirements (and a little more) are encompassed by the PERFICT principles for applied ecological modelling (McIntire et al. 2022).
It can be very challenging to fulfill these expectations, especially for non-programmers.
SpaDES aims to help (applied) ecologists “get there” by relieving ecologists from needing to know/learn advanced computer programming (e.g. developing caching mechanisms) and offering tools that will enable them to develop PERFICT workflows.
It does so by harnessing the flexibility of R, the contributions of its vast community and knowing what tools applied ecologists (and modellers) often need.
The SpaDES set of R packages is full of tools like prepInputs (Tip 1).
This book is intended to show applied ecologists (and whoever else!) the approaches that we use to bring our work closer to the PERFICT principles, better meet the expectations, and better address the problems.
Our challenge
With the limited training provided in this book, applied ecologists can work towards creating PERFICT workflows. Embracing the transparency and nimbleness it provides, we can pivot more rapidly to the current management needs. When we are asked at the end of a 3-year project, “can we redo everything because we have a new stakeholder who feels that our assumptions are wrong?”, our answer should be “yes”.