Code
# install/load necessary packages
repos <- c("predictiveecology.r-universe.dev", getOption("repos"))
install.packages(c("SpaDES.project", "Require"), repos = repos, dependencies = TRUE)
library(SpaDES.project)setupProjectCeres Barros
July 3, 2024
See Barebones R script for the code shown in this chapter
Castor is a forest and land-use model used to simulate forest harvest and its effects on multiple forest values, which include not only timber, but also habitat for several wildlife (e.g. caribou, fisher). It is a fully open-source model, implemented in SpaDES, developed and maintained by researchers at the Forest Analysis and Inventory Branch, BC Ministry of Forests.
In this chapter, we demonstrate how to set up a Castor workflow using setupProject from the SpaDES.project package. The code was adapted from this Castor scenario, with some modifications to streamline the code and accommodate the use of SpaDES.project functions.
A bare-bones version of this example is also available in this .R script
Currently, SpaDES.project functions expect SpaDES modules to either exist locally, or at root level of separate repositories. This is not the case with Castor modules, which are in a nested folder (i.e. a folder in a folder tree) of a single GitHub repository.
SpaDES.project will eventually be able to deal with this1, but for now we work around this issue by using a beta version from the nestedGHmodules branch.
setupProject does all the heavy-lifting of setting up the workflow, creating all the necessary folders, namely installing all package dependencies listed in the several modules and in setupProject(..., packages), sourcing additional files and creating parameter, inputs and outputs lists – see ?setupProject.
Note that all folder/file paths are relative to projectPath. Note also how setupProject is getting extra functions and the parameters list from online .R scripts that it downloads to the project folder and sources2.
setupProject(..., Restart = TRUE)
When creating a project for the first time, setupProject() will not only create the project folder structure, but also create an RStudio project file (.Rproj) and copy the current script (if it has been saved) into the project root directory.
Setting Restart = TRUE will open a new RStudio window from “inside” the project being defined (i.e., the RStudio Project will be opened and all paths will be relative to the project root directory) and open the active script.
This means that all code before the setupProject() call and the setupProject() call will need to be re-run again on the new RStudio/R sessions.
None of this happens if the RStudio project file is already active (open).
# set up the workflow paths, dependencies and modules
# as well as simulation parameters, (some) inputs and outputs
out <- setupProject(
paths = list("inputPath" = "modules/forestryCastor/inputs",
"outputPath" = "R/scenarios/comparison_stsm/outputs",
"modulePath" = "modules/",
"cachePath" = "modules/forestryCastor",
"projectPath" = "~/SpaDES_book/castorExample"),
modules = file.path("bcgov/castor@main/R/SpaDES-modules",
c("dataCastor",
"growingStockCastor",
"blockingCastor",
"forestryCastor",
"roadCastor")),
options = list(repos = repos),
functions = "bcgov/castor@main/R/functions/R_Postgres.R",
# install and load
require = "dplyr",
# install but don't load these:
packages = c(
"DBI",
"DiagrammeR",
"keyring",
"rgdal",
"RPostgreSQL",
"sp",
"terra"
),
params = "PredictiveEcology/PredictiveEcology.org@main/tutos/castorExample/params.R",
times = list(start = 0, end = 20),
outputs = {
data.frame(objectName = c("harvestReport",
"growingStockReport"))
},
scenario = {
data.table(name = "stsm_base_case",
description = paste("Priority queue = oldest first. Adjacency constraint",
"= None. Includes roads (mst) and blocks (pre).",
"Harvest flow = 147,300 m3/year in decade 1, 133,500",
"m3/year in decade 2, 132,300 m3/year in decades 3 to",
"14 and 135,400 m3/year in decades 15 to 25.",
"Minimum harvest age = 80 and minimum harvest volume = 150"))
},
harvestFlow = {
rbindlist(list(data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 1,
to = 1,
by = 1),
1),
flow = 1473000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 2,
to = 2,
by = 1),
1),
flow = 1335000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 3,
to = 14,
by = 1),
1),
flow = 1323000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 15,
to = 25,
by = 1),
1),
flow = 1354000,
partition_type = 'live')
))
},
# overwrite = TRUE, ## activate if you want to keep modules up-to-date the with online repository
Restart = TRUE
)simListsetupProject() returns a names list containing values that can be passed as argument to simInit()3.
We use do.call() to pass the whole list of arguments to simInit.
Another (more verbose) option would to call simInit directly:
Use the following functions to access workflow/model properties. events(), for instance will output the scheduled events, which at this point are only the init events of each module as you can see in the output below.
eventTime moduleName eventType eventPriority
<num> <char> <char> <num>
1: 0 checkpoint init 0
2: 0 save init 0
3: 0 progress init 0
4: 0 load init 0
5: 0 dataCastor init 1
6: 0 growingStockCastor init 1
7: 0 forestryCastor init 1
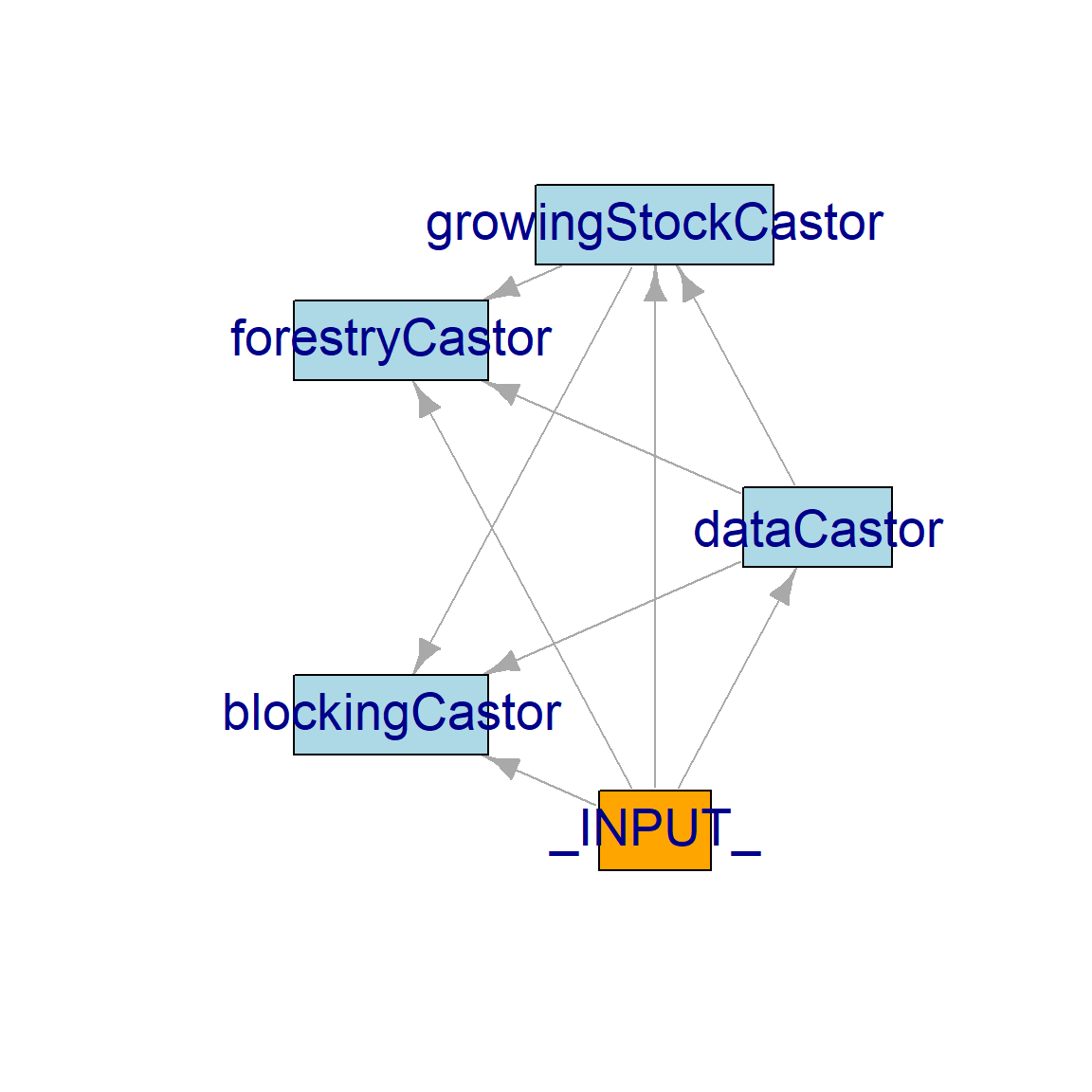
8: 0 blockingCastor init 1moduleDiagram() and objectDiagram() are great to visualise how each module interacts with the other. Recall that these interactions arise from object “exchanges” between modules, which are deduced by simInit() from module metadata (Figure 16.2) – i.e., if a module’s inputs are another’s outputs, then the first module will follow the second.

spades() runs the simulation, beginning with the execution of the init events. Notice how the result of outputs() differs from previously.
objectName
1 harvestReport
2 growingStockReport
file
1 C:/Users/cbarros/Documents/tutos/castorExample/R/scenarios/comparison_stsm/outputs/harvestReport_year20.rds
2 C:/Users/cbarros/Documents/tutos/castorExample/R/scenarios/comparison_stsm/outputs/growingStockReport_year20.rds
fun package saveTime saved arguments
1 saveRDS base 20 TRUE NA
2 saveRDS base 20 TRUE NAcompleted(castorSim) shows the chaining of events that was produced and run by spades(). The sequence of steps in the workflow therefore arises from each module’s events and their scheduling, rather than being explicitly imposed by the user.
eventTime moduleName eventType eventPriority
<num> <char> <char> <num>
1: 0 checkpoint init 0
2: 0 save init 0
3: 0 progress init 0
4: 0 load init 0
5: 0 dataCastor init 1
6: 0 growingStockCastor init 1
7: 0 forestryCastor init 1
8: 0 blockingCastor init 1
....We suggest omitting the blockingCastor module in setupProject() and rerunning the workflow again to see how spades is capable of re-generating a new workflow with little effort from the user.
modules <- c("dataCastor",
"growingStockCastor",
"forestryCastor")
out <- setupProject(
paths = list("inputPath" = "modules/forestryCastor/inputs",
"outputPath" = "/R/scenarios/comparison_stsm/outputs",
"modulePath" = "modules/",
"cachePath" = "modules/forestryCastor",
"projectPath" = "~/SpaDES_book/castorExample/"),
modules = modules,
functions = "bcgov/castor@main/R/functions/R_Postgres.R",
# install and load
require = "dplyr",
# install but don't load these:
packages = c(
"DBI",
"keyring",
"rgdal",
"RPostgreSQL",
"sp",
"terra"
),
params = "params.R",
times = list(start = 0, end = 20),
outputs = {
data.frame(objectName = c("harvestReport",
"growingStockReport"))
},
scenario = {
data.table(name = "stsm_base_case",
description = paste("Priority queue = oldest first. Adjacency constraint",
"= None. Includes roads (mst) and blocks (pre).",
"Harvest flow = 147,300 m3/year in decade 1, 133,500",
"m3/year in decade 2, 132,300 m3/year in decades 3 to",
"14 and 135,400 m3/year in decades 15 to 25.",
"Minimum harvest age = 80 and minimum harvest volume = 150"))
},
harvestFlow = {
rbindlist(list(data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 1,
to = 1,
by = 1),
1),
flow = 1473000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 2,
to = 2,
by = 1),
1),
flow = 1335000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 3,
to = 14,
by = 1),
1),
flow = 1323000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 15,
to = 25,
by = 1),
1),
flow = 1354000,
partition_type = 'live')
))
},
Restart = TRUE
)
# initialize and run simulation in one go
castorSim2 <- do.call(SpaDES.core::simInitAndSpades, out)# install/load necessary packages
repos <- c("predictiveecology.r-universe.dev", getOption("repos"))
install.packages(c("SpaDES.project", "Require"), repos = repos, dependencies = TRUE)
library(SpaDES.project)
# set up the workflow paths, dependencies and modules
# as well as simulation parameters, (some) inputs and outputs
out <- setupProject(
paths = list("inputPath" = "modules/forestryCastor/inputs",
"outputPath" = "R/scenarios/comparison_stsm/outputs",
"modulePath" = "modules/",
"cachePath" = "modules/forestryCastor",
"projectPath" = "~/SpaDES_book/castorExample"),
modules = file.path("bcgov/castor@main/R/SpaDES-modules",
c("dataCastor",
"growingStockCastor",
"blockingCastor",
"forestryCastor",
"roadCastor")),
options = list(repos = repos),
functions = "bcgov/castor@main/R/functions/R_Postgres.R",
# install and load
require = "dplyr",
# install but don't load these:
packages = c(
"DBI",
"DiagrammeR",
"keyring",
"rgdal",
"RPostgreSQL",
"sp",
"terra"
),
params = "PredictiveEcology/PredictiveEcology.org@main/tutos/castorExample/params.R",
times = list(start = 0, end = 20),
outputs = {
data.frame(objectName = c("harvestReport",
"growingStockReport"))
},
scenario = {
data.table(name = "stsm_base_case",
description = paste("Priority queue = oldest first. Adjacency constraint",
"= None. Includes roads (mst) and blocks (pre).",
"Harvest flow = 147,300 m3/year in decade 1, 133,500",
"m3/year in decade 2, 132,300 m3/year in decades 3 to",
"14 and 135,400 m3/year in decades 15 to 25.",
"Minimum harvest age = 80 and minimum harvest volume = 150"))
},
harvestFlow = {
rbindlist(list(data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 1,
to = 1,
by = 1),
1),
flow = 1473000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 2,
to = 2,
by = 1),
1),
flow = 1335000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 3,
to = 14,
by = 1),
1),
flow = 1323000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 15,
to = 25,
by = 1),
1),
flow = 1354000,
partition_type = 'live')
))
},
# overwrite = TRUE, ## activate if you want to keep modules up-to-date the with online repository
Restart = TRUE
)
# initialize simulation
castorInit <- do.call(SpaDES.core::simInit, out)
castorInit <- SpaDES.core::simInit(
times = out$times,
params = out$params,
modules = out$modules,
objects = list(scenario = out$scenario,
harvestFlow = out$harvestFlow)
)
# inspect the `simList`
SpaDES.core::params(castorInit)
SpaDES.core::inputs(castorInit)
SpaDES.core::outputs(castorInit)
SpaDES.core::times(castorInit)
# scheduled events
SpaDES.core::events(castorInit)
SpaDES.core::moduleDiagram(castorInit)
SpaDES.core::objectDiagram(castorInit)
castorSim <- SpaDES.core::spades(castorInit)
# we now have outputs
SpaDES.core::outputs(castorSim)
SpaDES.core::completed(castorSim)
modules <- c("dataCastor",
"growingStockCastor",
"forestryCastor")
out <- setupProject(
paths = list("inputPath" = "modules/forestryCastor/inputs",
"outputPath" = "/R/scenarios/comparison_stsm/outputs",
"modulePath" = "modules/",
"cachePath" = "modules/forestryCastor",
"projectPath" = "~/SpaDES_book/castorExample/"),
modules = modules,
functions = "bcgov/castor@main/R/functions/R_Postgres.R",
# install and load
require = "dplyr",
# install but don't load these:
packages = c(
"DBI",
"keyring",
"rgdal",
"RPostgreSQL",
"sp",
"terra"
),
params = "params.R",
times = list(start = 0, end = 20),
outputs = {
data.frame(objectName = c("harvestReport",
"growingStockReport"))
},
scenario = {
data.table(name = "stsm_base_case",
description = paste("Priority queue = oldest first. Adjacency constraint",
"= None. Includes roads (mst) and blocks (pre).",
"Harvest flow = 147,300 m3/year in decade 1, 133,500",
"m3/year in decade 2, 132,300 m3/year in decades 3 to",
"14 and 135,400 m3/year in decades 15 to 25.",
"Minimum harvest age = 80 and minimum harvest volume = 150"))
},
harvestFlow = {
rbindlist(list(data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 1,
to = 1,
by = 1),
1),
flow = 1473000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 2,
to = 2,
by = 1),
1),
flow = 1335000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 3,
to = 14,
by = 1),
1),
flow = 1323000,
partition_type = 'live'),
data.table(compartment = "tsa99",
partition = ' age > 79 AND vol > 149 ',
period = rep( seq (from = 15,
to = 25,
by = 1),
1),
flow = 1354000,
partition_type = 'live')
))
},
Restart = TRUE
)
# initialize and run simulation in one go
castorSim2 <- do.call(SpaDES.core::simInitAndSpades, out)SpaDES.project is currently being adapted to deal with modules nested in folders of GitHub repositories (instead of living in their own GitHub repositories). Hence, the code in this example is subject to changes in the near future.↩︎
Note that these files are placed in folders that respect the folder structure of where they come from (if they are not in the repository root folder). For instance, in the example the params.R is placed in <projectPath>/tutos/castorExample/params.R .↩︎